Figure 2.
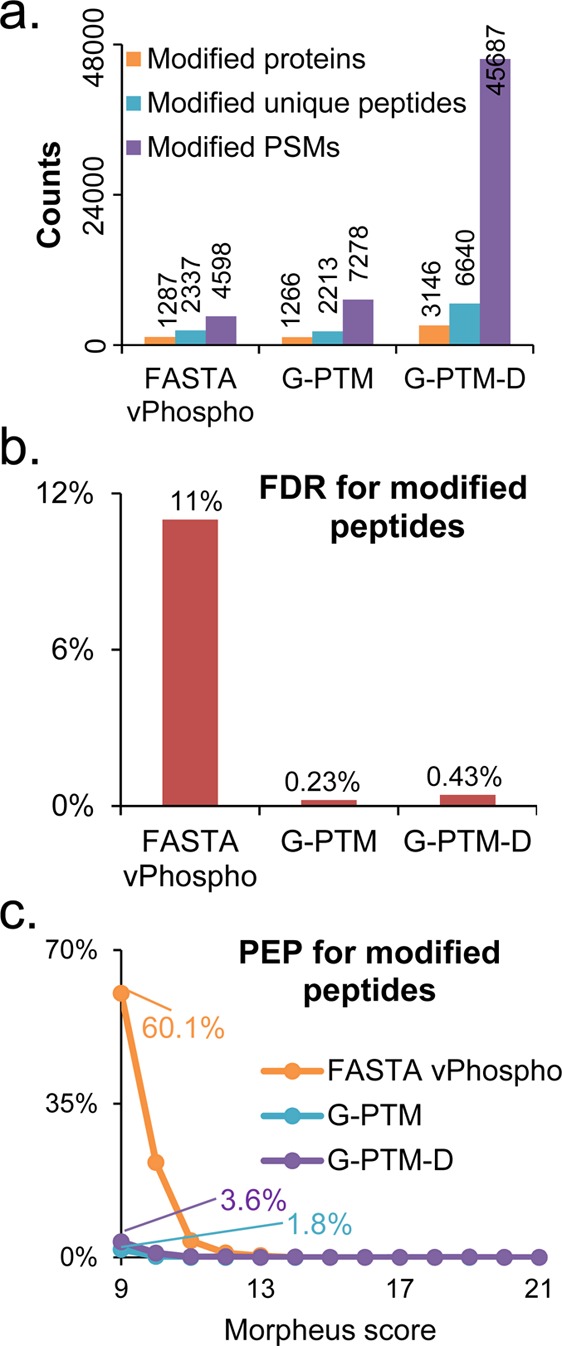
Results from three types of searches of the Jurkat cell data set: a vPhospho search (using the UniProt FASTA database with phosphorylation as a variable modification), a G-PTM search (using the PTM-curated UniProt database), and a G-PTM-D search. (a) Numbers of modified proteins, unique peptides, and PSMs for each search. The 45 687 modified PSMs identified by G-PTM-D are shown in Supplementary Table S2, with a hyperlink to the MS-Viewer report for each PSM. (b) False discovery rate (FDR) for modified peptides. (c) Posterior error probability (PEP) for modified peptides as a function of the Morpheus score. All results are based on 1% global FDR.
