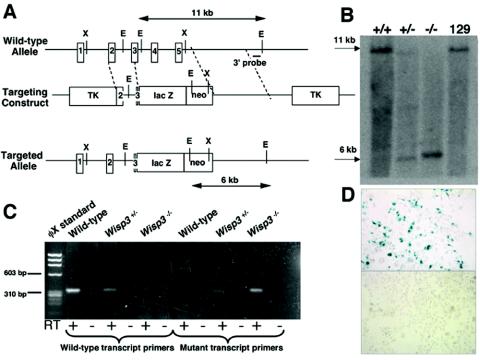
FIG. 2.
Targeted disruption of Wisp3 in mice. (A) Structure of the wild-type allele, the targeting construct, and the targeted allele (Wisp3-lacZ). EcoRI restriction enzyme sites (E) and restriction fragment lengths are noted. Black outlined boxes indicate individual exons. (B) Southern blot analysis of EcoRI-digested genomic DNA from control 129/SvEv mice and from Wisp3+/+, Wisp3+/−, and Wisp3−/− offspring of Wisp3+/− parents. Wild-type Wisp3 alleles are 11 kb and mutant alleles are 6 kb. (C) RT-PCR amplimers of wild-type and Wisp3-lacZ (mutant) transcripts from Wisp3+/+ (wild-type), Wisp3+/−, and Wisp3−/− mice. Wild-type transcript primers yielded the expected 311-bp product from wild-type and Wisp3+/− cartilage, but not from Wisp3−/− Wisp3−/− cartilage. Conversely, amplification with Wisp3-lacZ mutant transcript primers yielded the expected 308-bp product from Wisp3+/− and Wisp3−/− cartilage but not from wild-type cartilage. No amplimers were observed when non-reverse-transcribed RNA (RT −) was used as PCR template. (D) Photomicrographs (magnification, ×400) of Cos-7 cells transiently transfected with a vector expressing the Wisp3-lacZ fusion protein exhibit β-galactosidase activity (top), while nontransfected cells do not (bottom).