Figure 3.
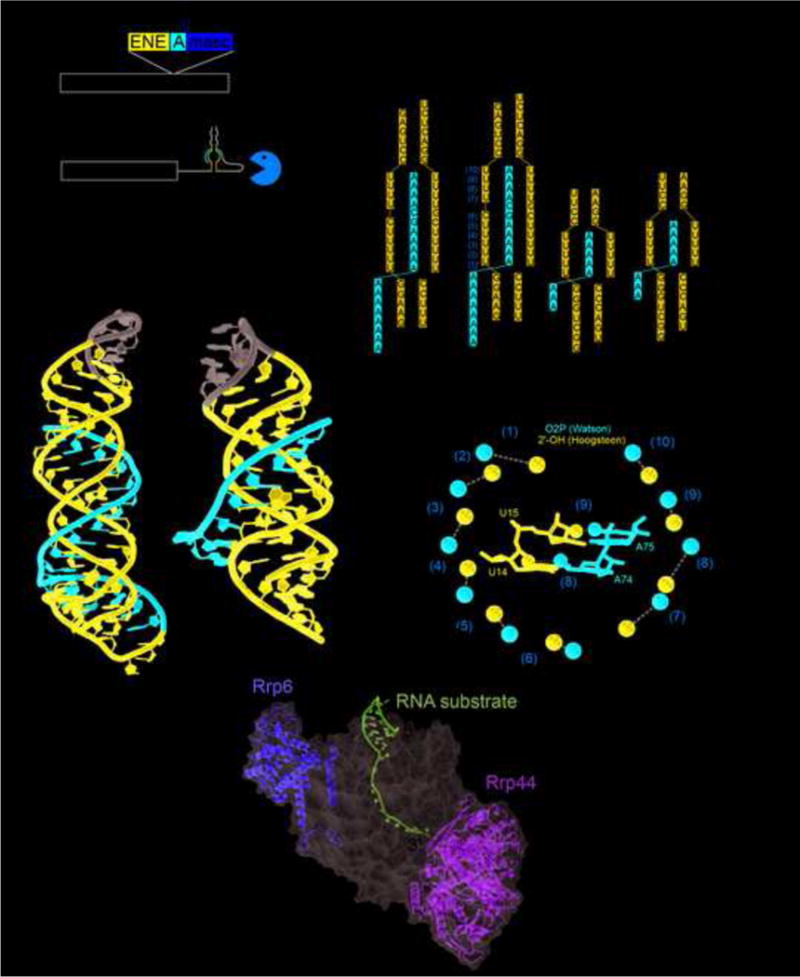
MALAT1 and PAN ENE crystal structures. (a) Schematic of the human MALAT1 transcript and the product after processing by RNase P (cut site indicated by arrowhead at nucleotide 8356). The exosome is able to degrade the unprocessed RNA, but the exosome is significantly hindered once the MALAT1 structure is at the 3′ end. (b) The crystal structure of MALAT1 (PDB: 4PLX) and PAN ENE (PDB: 3P22). The Watson strand with the 3′ end is in red, the Crick and Hoogsteen strands are in blue, and the engineered tetraloops are in gray. (c) The secondary structure of the conserved sequences and constructs crystallized for MALAT1 and PAN ENE. Leontis and Westhof symbols indicate non-WC base pairs. (d) Crystal structure of the 12-subunit exosome; a bound structured RNA with the 3′ end extending into the exosome is shown in magenta. Nucleases Rrp6 and Rrp44 are yellow and green, and the rest of the exosome is represented as a semi-transparent gray surface. (PDB: 5C0X). (e) The OP2 and 2′ OH of the Watson and Hoogsteen strands are shown as red or blue spheres respectively. The distances between them are given and orange numbering corresponds to the secondary structure in panel (b). The grey arrow depicts moving along the Watson strand from the 5′ to the 3′ direction and the location of the G-C pair in the middle that provides the “reset.” In the middle the same coloring is used to highlight interactions of the U14 and A74 (pair (8)) and U15 and A75 (pair (9)).
