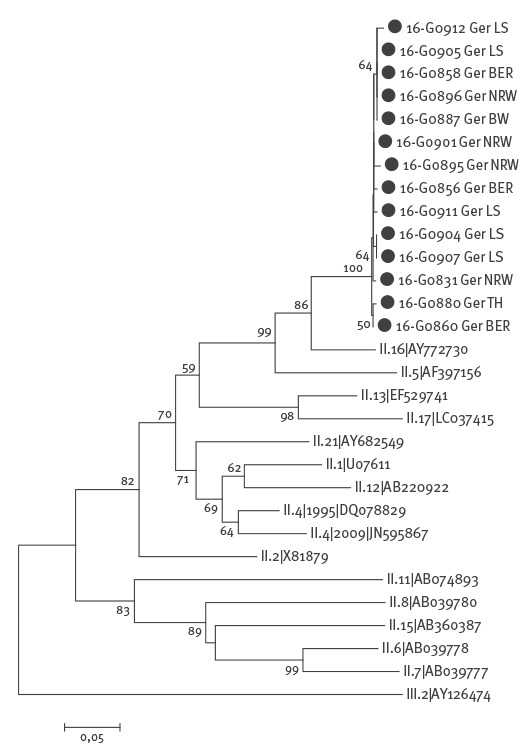
Figure 2.
Phylogenetic analysis based on the nucleotide sequence of a 357 bp region (ORF1) of genogroup II norovirus, Germany, 2016/17 (n = 14 representative samples)
BER: Berlin; BW: Baden-Wuerttemberg; Ger: Germany; LS: Lower Saxony; NRW: North Rhine-Westphalia; TH: Thuringia.
Analysed sequence: nucleotide positions 4,332–4,689 according to accession no: AY772730. Strains analysed in this study are denoted with a black bullet (•). Scale bar indicates nucleotide substitution per site. Sequence alignments were performed with the ClustalW algorithm. Neighbour-joining phylogenetic tree was produced using the MEGA 7 software with bootstrap test (1,000 replicates). Bootstrap values (1,000 replicates) above 50 are shown. The evolutionary distances were computed using the Kimura-2 parameter method.