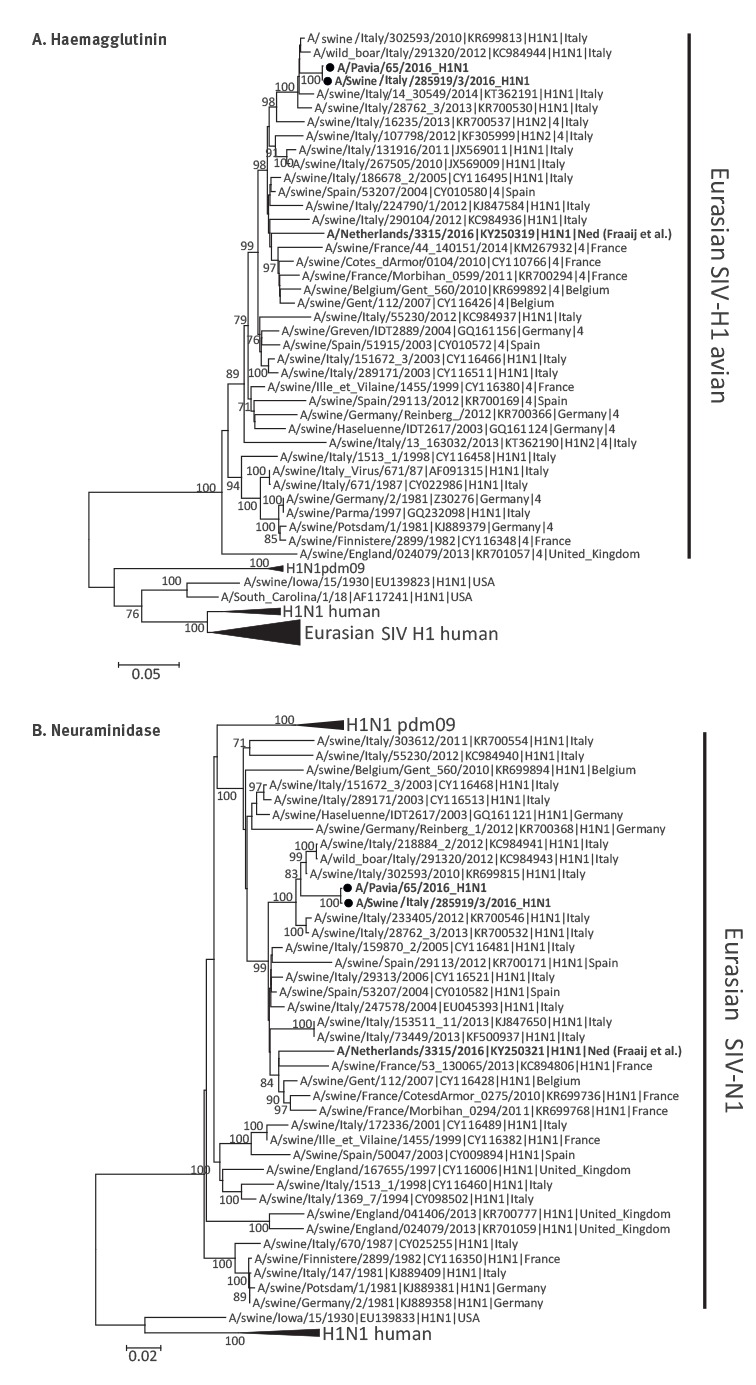
Figure 2.
Phylogenetic relationship of human and swine influenza A(H1N1) virus strains based on complete haemagglutinin and neuraminidase nucleotide sequences, compared with patient isolate, Italy, October 2016
SIV: swine influenza virus.
The unrooted trees were created by neighbour-joining method implemented in the MEGA 6.0 software and bootstrapped with 1,000 replicates. The pairwise distances were computed using the Kimura 2-parameter model. Only bootstrap values higher than 70% are shown. Patient and pig haemagglutinin and neuraminidase sequences from our study are marked with a black circle and bold letters. Scale bars indicate nucleotide substitutions per site. Sequence reported in Fraaij et al. [6] are highlighted in bold. The other sequences used in the phylogenetic trees were obtained from Watson et al. [12] as well as from the GenBank database.