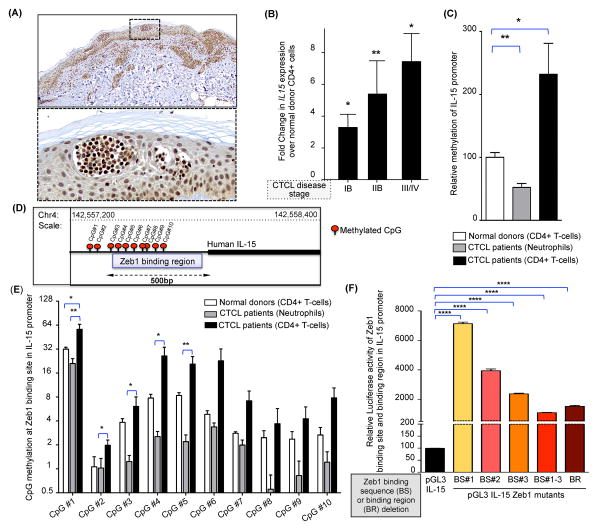
Figure 1. Overexpression of IL-15 in CTCL patient samples.
(A) Representative microscopic images of IL-15 immunohistochemical staining of a skin lesion from a CTCL patient. Scale bar=100μm. Dotted box in the upper panel indicates higher magnification of skin lesion presented in the lower panel. (B) Fold change in IL-15 transcript (mean ± SEM) in CD4+ T cells obtained from blood of patients with progressive stages of CTCL (N=3 each), relative to CD4+ T-cells in normal donor blood (N=3). IL-15 transcript was normalized to 18S and the values for normal donors were arbitrarily set at 1. (C) Graphical representation of IL-15 promoter methylation as determined by pyrosequencing in DNA extracted from sorted CD4+ T-cells and neutrophils of CTCL patients listed in Supplementary Table I. The relative quantity of promoter methylation was compared to purified CD4+ T-cells from normal donors, which is arbitrarily set at 100%. Data shown are mean ± SEM, N=9 for CTCL patients and N=6 for normal donors. (D) Diagram of regulatory region within the human IL-15 promoter, illustrating both the location of the putative Zeb1 binding sites within the CpG rich region and the extent of CpG methylation within this region (CpG1-CpG10) for a typical CTCL patient. (E) Differential methylation of CpG dinucleotides 1 through 10 with in a CpG rich Zeb1 binding region of the IL-15 promoter in CTCL patients (CD4+ T-cells and neutrophils) vs. normal donor CD4+ T-cells; data presented as mean ± SEM, N=9 for patients and N=6 for normal donors. (F) To characterize the transcriptional competence of Zeb1 binding site in IL-15 promoter, the pGL3-IL-15 plasmid construct (presented here as native promoter) was subjected to site directed mutagenesis to create IL-15 promoters lacking Zeb1 binding sites (BS#1, BS#2, BS#3, BS#1–3) or the entire binding region (BR). Relative luciferase activity was measured and normalized to a promoterless PGL3 basic vector (mean ± SEM, N=3 each). For each graphical representation in Figure 1, data are presented as mean ± SEM, *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001 unpaired or paired two-tailed student’s t-test.