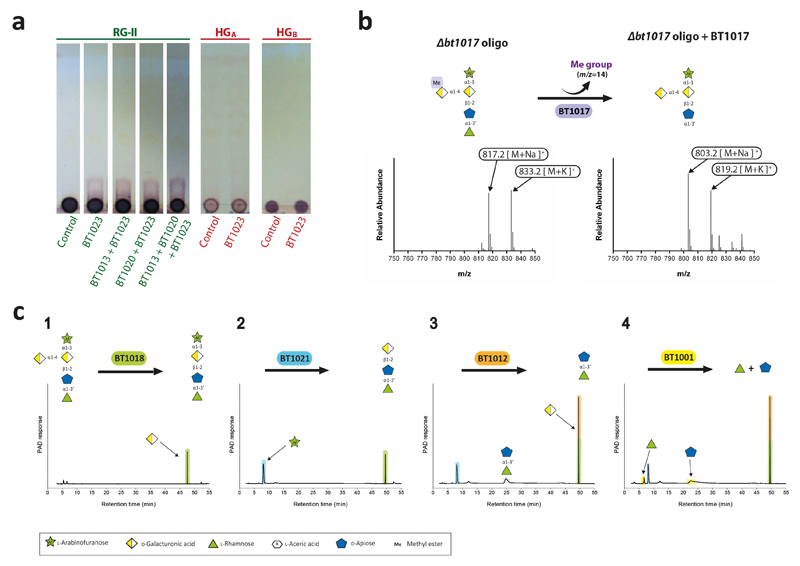
Extended Data Fig. 8. Identification of the enzymes that disassemble the backbone of RGII.
a, BT1023 was incubated with RG-II or homogalacturonan (1% w/v) in 50 mM CAPSO buffer, pH 9.0, containing 2 mM CaCl2. RG-II substrate was either untreated or had been previously incubated with BT1013 and/or BT1020. The reactions were analysed by TLC. Lanes labelled “control” were the appropriate glycan incubated with the appropriate buffer but without the inclusion of enzyme. b, the oligosaccharide generated by the Δbt1017 mutant (Δbt1017 oligo) was incubated with BT1017 and the reaction product was analysed by mass spectrometry. c,sequential degradation of the product generated in b by the enzymes indicated under standard conditions. The sugars released by these other enzymes (reactions 1 to 4) were identified and quantified by HPAEC-PAD. The structure of the oligosaccharide sugars followed the notation described in Fig. 1 and Extended data Fig. 2. The example shown are from technical replicates n = 3.