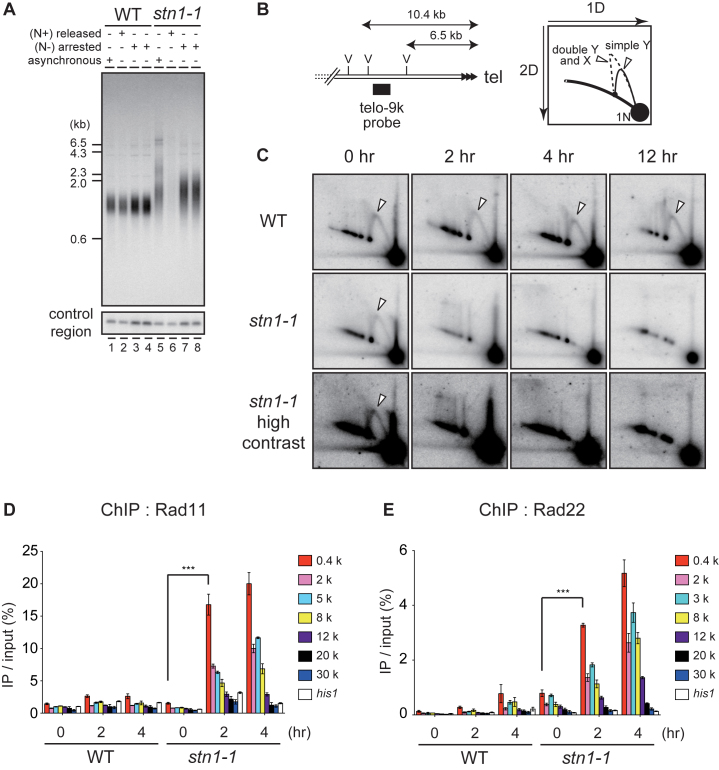
Figure 4.
stn1-1 at 36°C frequently arrests DNA replication at subtelomeres, leading to DSBs. (A) Southern hybridization was performed with EcoRI-digested genomic DNA from cells harvested after culture in the following conditions: asynchronously growing cells at 25°C in MM medium (lanes 1 and 5); starved for nitrogen for 16 h at 25°C and released by NH4Cl addition and cultured at 36°C for 24 h (lanes 2 and 6); starved for nitrogen for 16 h at 25°C then shifted to 36°C for 12 h (lanes 3 and 7) and 24 h (lanes 4 and 8), respectively. (B) Positions of the telo-9k probe and EcoRV sites (left). (C) Wild-type and stn1-1 were cultured at 25°C and 36°C for the indicated times. EcoRV-digests were resolved by neutral-neutral 2D gel electrophoresis and probed by Southern hybridization with the telo-9k probe. The white triangles indicate the replication intermediates, simple Y-arcs. (D) Wild-type and stn1-1 cells expressing C-terminally myc-tagged Rad11 were cultured at 36°C for the indicated times and were analyzed by ChIP-qPCR using qPCR primer sets depicted in Figure 2A. Error bars show mean values of three independent experiments with SD. *** P value was 0.0043 calculated with a two-tailed Student's t-test. (E) Wild-type and stn1-1 cells expressing C-terminally myc-tagged Rad22, cultured at 36°C for the indicated times, were analyzed by ChIP-qPCR. Error bars show mean values of three independent experiments with SD. *** P value was 0.0018 calculated with a two-tailed Student's t-test.