Abstract
Ribozymes can catalyze phosphoryl or nucleotidyl transfer onto ribose hydroxyls of RNA chains. We report a single ribozyme that performs both reactions, with a nucleobase serving as initial acceptor moiety. This unprecedented combined reaction was revealed while investigating potential contributions of ribose hydroxyls to catalysis by kinase ribozyme K28. For a 58nt, cis-acting form of K28, each nucleotide could be replaced with the corresponding 2΄F analog without loss of activity, indicating that no particular 2΄OH is specifically required. Reactivities of two-stranded K28 variants with oligodeoxynucleotide acceptor strands devoid of any 2΄OH moieties implicate modification on an internal guanosine N-2, rather than a ribose hydroxyl. Product mass suggests formation of a GDP(S) adduct along with a second thiophosphorylation, implying that the ribozyme catalyzes both phosphoryl and nucleotidyl transfers. This is further supported by transfer of radiolabels into product from both α and γ phosphates of donor molecules. Furthermore, periodate reactivity of the final product signifies acquisition of a ribose sugar with an intact 2΄-3΄ vicinal diol. Neither nucleobase modification nor nucleotidyl transfer has previously been reported for a kinase ribozyme, making this a first-in-class ribozyme. Base-modifying ribozymes may have played important roles in early RNA world evolution by enhancing nucleic acid functions.
INTRODUCTION
In vitro evolution has established that nucleic acids can catalyze diverse chemical reactions (1–6). This versatility provides experimental support for key tenets of the RNA world hypothesis, which posits a major role for RNA enzymes (ribozymes) prior to the appearance of genetically encoded protein enzymes (6), and it sets the stage for exploiting ribozymes in bottom-up synthetic biology.
Phosphoryl transfer is a key reaction in many facets of biology, such as leaving group activation, compartmentalization of metabolites and signal transduction. Nucleic acid sequence space is densely populated with RNA and DNA molecules that catalyze phosphoryl or thiophosphoryl group transfer (2,7–14). All characterized kinase ribozymes and deoxyribozymes promote O-phosphorylation of a polynucleotide 5΄OH, 3΄OH or 2΄OH moiety (1,2,15,16). Kinase ribozymes could have supported complex small-molecule metabolism during an RNA world by promoting phosphorylation of numerous other molecules, although the potential of ribozymes to utilize alternative phosphoryl acceptors remains largely unexplored. In particular, there are no reports of nucleobase phosphorylation, N-phosphorylation of any substrate, or phosphorylation of amino acids or other small metabolites by nucleic acid catalysts, although deoxyribozymes that phosphorylate small peptides have been described (17). Hence, much of chemical space remains to be explored.
Covalently modified nucleobases, such as those found in tRNA and rRNA, can significantly augment the functional repertoire of nucleic acids. For example, in some bacterial tRNAs, 4-thio-uridine promotes crosslinking with cytosine in the presence of UV, rendering the tRNA as poor substrates for tRNA synthetases and ultimately turning on the stress response (10). In vitro selections have been performed using libraries that contained modified nucleobases that mimic amino acid side chains or provide other chemical functionalities. Among these, partially hydrophobic SOMAmer nucleic acids provide dramatic examples of how the chemical diversification of nucleobases enables the evolutionary exploration of new and unusual structures and binding modes (18–20).
The present study describes an unexpected intersection between these two themes. While investigating 2΄OH requirements for truncated forms of kinase ribozyme K28, we found that no specific 2΄OH is required in either the ribozyme or substrate strand, and that the acceptor is the nucleobase of an internal (deoxy)guanosine. Although this ribozyme was initially selected for self-thiophosphorylation, mass spectrometry revealed a much larger adduct that appears to be formed in a multi-step reaction that includes two non-equivalent (thio)phosphoryl transfers and addition of GMP. Nucleobase phosphorylation occurs during nucleotide biosynthesis and catabolism, but the nucleotidyl transfer that we observe is distinct from common biological nucleotidyl transfers, such as the formation of an mRNA 7-methylguanylate cap (21). Instead, analog reactivity and product decomposition in mild acid implicate N-phosphorylation on the guanosine Watson–Crick face. We have recently demonstrated that variants of this ribozyme can be used to selectively modify and regulate functional RNAs (22). Nucleobase modification by ribozymes provides an avenue to explore an expanded chemical space in RNA world scenarios, and the free 2΄ and 3΄ hydroxyls of the attached guanosine ribose also provide opportunities for chain elongation from this unusual branch point. Our observation of combined nucleobase thiophosphorylation and nucleotidyl transfer provides the first experimental support for RNA catalysis of both classes of reactions by a single ribozyme.
MATERIALS AND METHODS
Oligodeoxynucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and TriLink BioTechnologies (San Diego, CA). RNA was transcribed in vitro using wild type or Y639F mutant phage T7 RNA polymerase, which were overexpressed as His-tagged fusion proteins in bacteria and purified in the laboratory. 2΄F nucleotides were purchased from TriLink Biotechnologies (CA, USA) and Jena Bioscience (Jena, Germany). GTPγS was purchased from Sigma (St. Louis). Radiolabeled nucleotides [γ32P]-ATP, [α32P]-CTP and [α32P]-GTP, were purchased from Perkin-Elmer (Waltham, MA). Ribozymes were either transcribed using 33nM [α32P]-CTP or non-radioactive CTP followed by 5΄ end labeled using [γ32P]-ATP and PNK (NEB). Tri-layered organomercurial gels containing 100 μg/ml [(N-acryloylamino)phenyl]mercuric chloride (APM) chloride in the middle layer were prepared as described (23) to capture sulfur-containing polynucleotides formed by thiophosphoryl transfer from GTPγS.
Thiophosphorylation reaction
10 000–20 000 cpm of [5΄-32P] end-labeled substrate strand (50–166 nM) was added to 2 μM Ribozyme strand in water. Ribozyme:substrate mixture was heated at 85°C for 5 min. 5X Thiophosphorylation buffer (1X = 25 mM Tris•HCl pH 8.0, 30 mM MgCl2, 10 μM CuCl2, 200 mM KCl and 15 mM NaCl) was added and the tubes were incubated at room temperature for 5 min and moved to ice to form the ribozyme:substrate complex. Reactions (50 or 100 μL) were initiated by adding 1 mM GTPγS (final concentration). Tubes were then incubated at 32 °C for 6 h unless stated otherwise. An equal volume of 90% formamide, 50 mM EDTA was then added to quench the reaction. Products were separated on 10% denaturing tri-layered APM gels. Autoradiographs were obtained with a FLA-9000 GE phosphorimager (FujiFilm) and analyzed with MultiGauge software (Version 3.0). The kinetic data at pH 7.5 and 8.0 were fit to equation f(t) = fmax•(1 – e−kobs•t); for pH 7.0, data were fit to equation f(t) = fmax•kobs•t using the same fmax values as for pH 8.0.
Temperature and pH stability of thiophosphorylated product
Thiophosphorylated products were ethanol precipitated, resuspended in water, then added to tubes containing 30 mM MgCl2, 200 mM KCl, 15 mM NaCl and 25 mM of the appropriate pH buffer: Tris for pH 7.0 to 9.0, MOPS for pH 6.0 to 7.0, MES for pH 5.5 to 6.0, or acetate for pH 4.5–5.5. Products were separated on 10% denaturing tri-layered APM gels. For temperature stability experiments, thiophosphorylated products were ethanol precipitated and incubated in 25 mM Tris pH 8.0 buffer at the indicated temperatures.
Primer extension of thiophosphorylated product
Non-radiolabeled acceptor strands (d3.1 or 5΄-extended d3.1) were thiophosphorylated as above and purified from denaturing trilayer APM gels. These DNAs were annealed to 5΄-32P-labeled primers by heating to 95°C for 3 min, incubating at room temperature for 5 min and then moving them to ice. Reverse transcription reactions were initiated by the addition of 5 U/μl Moloney-Murine Leukemia Virus (M-MLV) reverse transcriptase (Promega) and incubated at 37°C in 50 mM Tris–HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2, 0.25 mM dNTPs (each) and 10 mM DTT for 20 min. Reactions were stopped by adding equal volume of 90% formamide and 50 mM EDTA and separated on a 15% denaturing PAGE.
Dye-labeling
Thiophosphorylation reactions were carried out for 6 h (∼30% yield) and were precipitated and resuspended in 10 mM NaIO4, followed by 2 h incubation at 4°C in the dark. HEPES pH 8.5 was added after 2 h to a final concentration of 100 mM. 3 μl (15 ng) of Cy-3 hydrazide (Lumiprobe, resuspended in 100% DMSO) was added to the sample. Reactions were incubated for 2 h at room temperature in the dark, followed by ethanol precipitation. Samples were resuspended in 95% formamide and 50 mM EDTA, and aliquots were separated by 20% denaturing PAGE.
[α-32P]-GTP reaction
Thiophosphorylation reactions were carried out as previously described using unlabeled substrate and ribozyme (1 μM each). Unlabeled GTP (indicated final concentration) was added and samples were incubated in ice for 5 min. [α32P]-GTP was added to a final concentration of 0.5 μM. Reactions were incubated at 10°C for 18 h and separated by 20% denaturing PAGE.
Mass spectrometry
Samples were prepared as described by Koomen et al. (24). In brief, thiophosphorylation reactions were carried out as above, and products were ethanol precipitated using ammonium acetate. Pellets were dried and either resuspended in water or first resuspended in 100 mM acetate buffer, pH 4.5, before being incubated at 37°C for 5 h. Samples were precipitated again using ammonium acetate and resuspended in water. For the ‘no reaction’ control, the d3.1 substrate was resuspended in thiophosphorylation buffer, and precipitated twice as above. Polynucleotide kinase (PNK) control was generated by thiophosphorylation of the substrate using PNK and ATPγS as the donor, then precipitated twice as above. All samples were resuspended in water at 5–7 μM, then treated with Dowex resin charged with ammonium citrate and mixed with equal volumes of diammonium citrate (10 mg/ml) and 2,3,4-trihydroxyacetophenone (THAP) (16 mg/ml in methanol) to make the spotting solution, which was spotted on THAP crystals. Mass spectrometry was performed on an AB Sciex Voyager DE MALDI TOF mass spectrometer at the Proteomics Core of the University of Missouri.
RESULTS
2΄Fluoro substitutions do not inhibit ribozyme activity
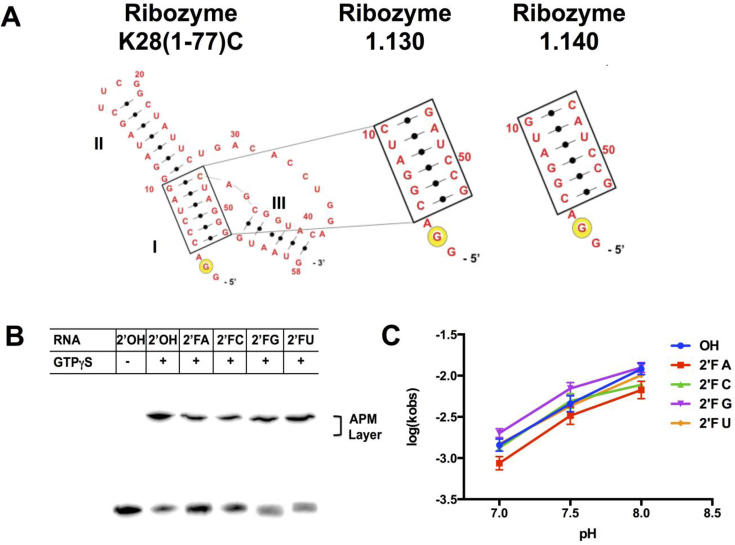
Ribozymes K28(1-77)C, 1.130 and 1.140 are truncated forms of the Cu2+ dependent ribozyme K28 (25,26), which was initially selected for self-thiophosphorylation (1). For ribozyme 1.130, five base pairs in stem I are flipped to their Watson–Crick complement relative to K28(1-77)C, while in ribozyme 1.140, four base pairs are flipped (Figure 1A). These changes increase product yield (26) and disrupt a palindrome in the original sequence that otherwise inhibits K28(1-77)C at high RNA concentrations (Supplementary Figure S1). To explore 2΄OH requirements in thiophosphoryl transfer, ribozyme 1.140 was transcribed either with 2΄OH at every position or with one of the four NTPs completely substituted with its 2΄F-NTP analog. In general, RNA and 2΄F-RNA duplexes are very similar (27), but ligand binding and catalysis can be directly influenced by how a given 2΄ moiety is utilized. After incubation with GTPγS and γ-thiophosphoryl transfer onto the RNA, product retention in the organomercurial layer (APM) of a tri-layer gel was observed for all five versions of the ribozyme, signifying that all five are catalytically active (Figure 1B). Product yields were equivalent or only modestly reduced (all values within 25% of the mean) across the set in the order 2΄OH > 2΄F-U ≈ 2΄F-G > 2΄F-A ≈ 2΄F-C. Thus, each of the four nucleotides can be uniformly substituted with 2΄F. Ribozymes transcribed with multiple 2΄F-NTPs were less active (Supplementary Figure S2A), indicating that the 2΄OH groups for at least two NTP sets need to be substituted to 2΄F for inhibition to occur. The rate of product formation increased with pH for all five mono-substituted species (Figure 1C), and product yield was greatly diminished in the absence of Cu2+ (Supplementary Figure S2B), as observed previously for ribozyme K28(1–77)C (25). These data indicate that neither the pH nor metal ion dependence of the reaction derives from any one specific 2΄OH, and that 2΄F substitution does not markedly change the chemical activity of the ribozyme.
Figure 1.
2΄F-substituted ribozymes are active. (A) Secondary structures of ribozymes K28 (1-77)C, 1.130 and 1.140. Stems I, II and III are indicated, and the major site of self-phosphorylation is shown with the yellow circle. (B) Transcripts of ribozyme 1.140 containing either all 2΄OH or complete 2΄F substitutions on the indicated nucleotides were allowed to self-thiophosphorylate for 6 hr. Samples were separated on tri-layered organomercurial (APM) gels. Product yields for 2΄OH, 2΄F-A, 2΄F-C, 2΄F-G and 2΄F-U were 74 ± 3%, 49 ± 7%, 50 ± 5%, 59 ± 4% and 67 ± 8%, respectively for three independent experiments. (C) Thiophosphorylation reactions using 2’F substituted RNAs were performed at pH 7.0, 7.5 and 8.0, and single-turnover rate constants were calculated as described in Methods. Error bars represent standard error of mean from at least three independent measurements.
Phosphoryl acceptor is a guanine nucleobase
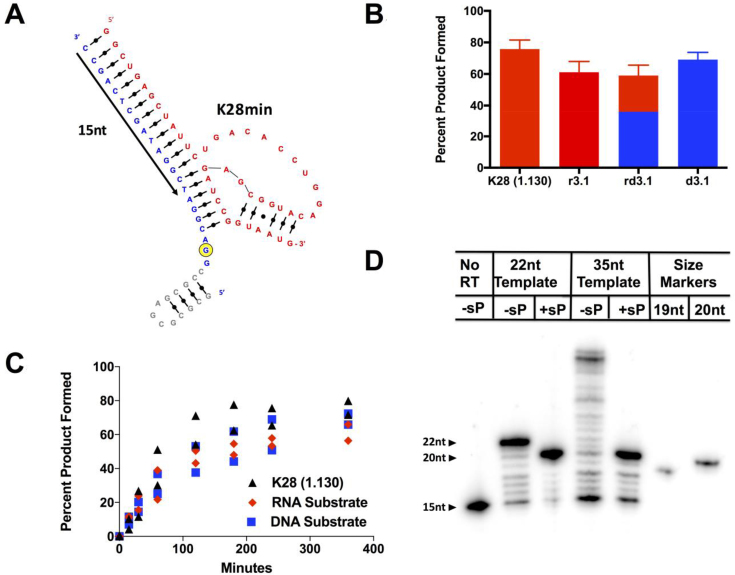
The near-normal activity in the 2΄F-substituted ribozymes could be explained by the acceptor being a species other than a ribose hydroxyl, which would indicate that these RNAs catalyze a reaction that has not previously been observed for ribozymes. To explore this possibility, we generated a two-stranded version of the closely-related ribozyme 1.130 by annealing gel-purified catalytic strand (44nt, designated ‘K28min,’ Figure 2A, shown in red) with acceptor strands (22nt) through a 19nt internal guide sequence (IGS) in the catalytic strand. The first three chemically synthesized acceptors to be tested were an all-RNA substrate, an all-DNA substrate (Figure 2A, shown in blue), and an RNA/DNA chimera substrate with deoxy substitutions at positions 4 to 22. Annealed complexes were incubated with GTPγS, and reactions were evaluated on APM gels. Product yields and rate constants were essentially indistinguishable for all three trans-acting complexes, even when the substrate strand was composed entirely of DNA (Figure 2B), and they were similar to the corresponding values for ribozyme 1.130 (Figure 2C). The original ribozyme K28 and several smaller derivatives have been shown to transfer radiolabel from [γ-32P]GTP onto both a major site near the 5΄ end and a second, minor internal site (26). For the two-stranded design based on ribozyme 1.130, activity is observed only at the major site near the 5΄ end, perhaps due to sequence disruptions near the internal acceptor site (Supplementary Figure S3A), fortuitously simplifying analysis by removing potential complications that might arise from partial reactivity at the minor internal site.
Figure 2.
Phosphoryl acceptor is the nucleobase of dG2. (A) Trans-acting, two-stranded version of ribozyme 1.130. Catalytic strand (RNA) is shown in red, acceptor substrate strand (DNA) is shown in blue. Yellow circle denotes the major acceptor site. Nucleotides in grey indicate extra 13 nucleotides added to d3.1 to generate the 5΄-extended DNA substrate. Nucleotides spanned by the primer used in (D) are depicted by black arrow. (B) Percent product formed for the unimolecular, full-length ribozyme and by the trans-acting version of the ribozyme with different nucleic acid substrates. Red = RNA; blue = DNA; split color = chimera substrate. Plotted values are averages of 3 measurements, errors represent standard error of mean. (C) Product yield for the unimolecular ribozyme and for trans-acting ribozyme K28min with nucleic acid substrates r3.1 and d3.1 as a function of time (0, 15, 30, 60, 120, 180, 240 and 300 min). Fitting the data to a first-order kinetic equation yielded kobs values of 0.013 ± 0.002 min−1 for cis-acting ribozyme 1.130 and 0.013 ± 0.003 and 0.008 ± 0.002 min−1 for trans-acting ribozymes with RNA and DNA substrates, respectively. Uncertainties represent the range of values for 2 replicate experiments. (D) Primer extension on thiophosphorylated product results in a 20mer extended product with both d3.1 (22nt) and the 5΄-extended (35nt) substrates, consistent with strong polymerase blockage arising from dG2 modification.
To map the modification site that is produced by ribozyme K28min, reverse transcription pausing was monitored for acceptor strand d3.1 and for a variant acceptor in which the 5΄ end was extended by 13nt (Figure 2A, extension in gray). Both acceptor strands were incubated with ribozyme K28min and GTPγS, and the products were purified from a denaturing APM gel and annealed to a 5΄-radiolabeled primer (Figure 2A, black arrow). Primer extension by M-MLV reverse transcriptase (RT) produced a strong polymerization stop after position A3, irrespective of whether the 5΄ extension was present. These data rule out the prior interpretation (26) of RT termination at A3 as being due to known RT artifacts at RNA 5΄ ends (see Supplementary Text), and they rule out 5΄OH and 5΄PO4 as possible acceptor sites. Instead these results implicate position dG2 as the site of modification (Figure 2D). Because the only remaining major nucleophiles in this region are the heteroatoms of the nucleobase, these data also imply that this ribozyme catalyzes modification directly upon the nucleobase of dG2.
Acceptor analog substitutions at dG2 identify N-2 as the acceptor
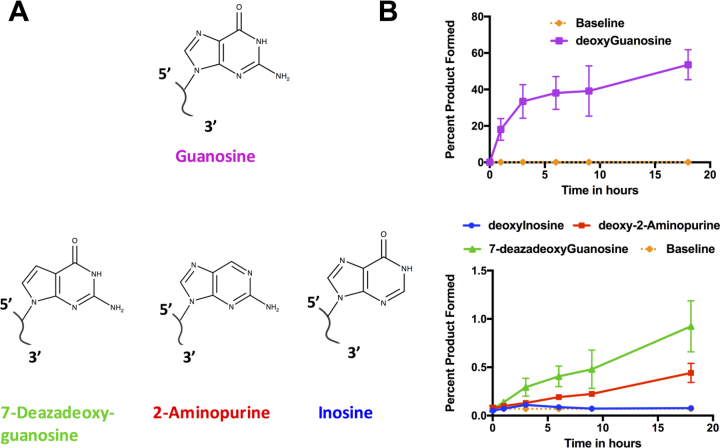
To identify the nucleophilic acceptor atom in dG2, we measured product yields for synthetic acceptor strand analogs annealed to ribozyme K28min. Low-level product accumulation was observed for acceptors carrying 2΄-deoxy-7-deazaG or 2΄-deoxy-2-aminopurine (2AP) substitutions in place of dG2, and these products continued to accumulate with time. These results rule out a requirement for N-7 or O-6. In contrast, no product was observed when dG2 was replaced with inosine (I), which retains O-6 but lacks the exocyclic N-2 amine (Figure 3 and Supplementary Figure S3B). Thus, N-2 is required for product formation and might act as the acceptor.
Figure 3.
Identification of guanosine N-2 as acceptor site. (A) Molecular structures of deoxyguanosine analogs used in acceptor substrate substitution analysis. (B) Quantification of product accumulation for these analogs. Thiophosphorylation reactions were carried out with DNA acceptor substrates carrying the indicated substitutions for dG2 and quantified on APM gels. Plotted values are means of at least two experiments. Percent product formed with deoxyguanosine (top) and different analogs (bottom) as the substrate. The orange baseline is the average of all the experiments at time 0, representing background retention at APM layer. Time points were taken after incubating 0, 1, 3, 6, 9 and 18 h at 10°C. Representative APM gels used in this quantification are shown in Supplementary Figure S3B.
To gain further insight into how the unpaired 5΄-d(GGA) trinucleotide is accommodated within the folded ribozyme, we next determined nucleotide requirements at the two positions flanking the dG2 acceptor within this segment. Replacing dG1 with canonical nucleotides (dA, dC or dT) or with dI or d2AP eliminated or severely impaired product formation at 32°C (Supplementary Figure S4A and S4B), but product formation was restored at 10°C for the dG1→dI substitution (Supplementary Figure S4C). Position dA3 was similarly sensitive to substitution with dC, dG or dT, but was more forgiving for the dA3→d2AP or dA3→dI substitutions (∼12%) (Supplementary Figure S4B). When the substrate contained a flexible hexaethylene glycol linker after the second or the third nucleotide (Supplementary Figure S5A), product yield increased as the temperature was reduced in most cases (Supplementary Figure S5B). No product was observed when 5΄d(GGA) was supplied in trans to a ribozyme lacking this triplet, potentially indicating weak interaction with the acceptor strand. To test this idea, we next explicitly evaluated the effect of temperature on product accumulation and stability. Using d3.1 as the acceptor strand, ribozyme K28min accumulated product much more rapidly at 10 or 20°C than at 30 or 40°C, and was essentially inactive at 50°C (Supplementary Figure S6A). However, once the product was generated and purified, it was stable against thermal degradation at temperatures below 50°C (Supplementary Figure S6B), ruling out product instability as an explanation for the low-temperature preference. Instead, the ribozyme΄s low-temperature optimum and its sensitivity even to small changes in the identity or entropy of flanking nucleotides likely indicate that 5΄d(GGA) is accommodated within the active site via weak, highly-specific tertiary interactions.
An unusual, RNA-catalyzed nucleotidyl transfer
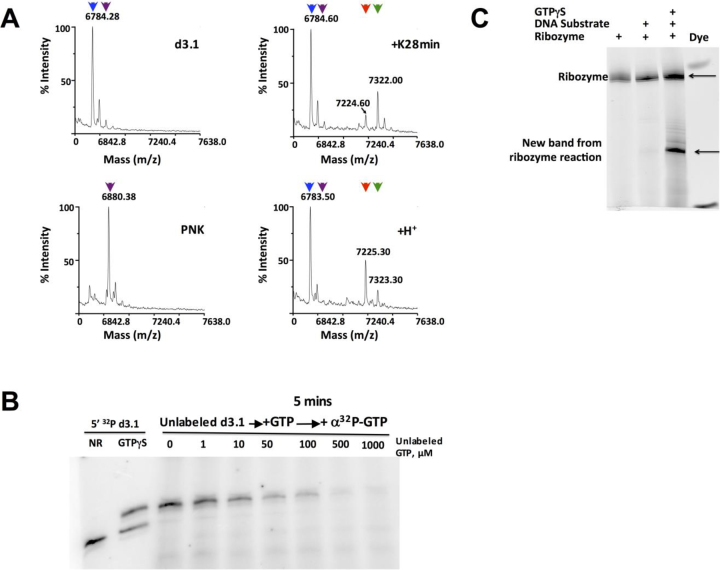
Mass spectrometry of purified product revealed a major peak that is shifted +537.4 m/z relative to unreacted substrate strand (Figure 4A top right). This is much greater than the shift associated with thiophosphorylation alone (+95.8 m/z, Figure 4A bottom left) and instead corresponds almost exactly to the mass shift calculated for addition of both a thiophosphoryl group and a sulfur-containing nucleotide diphosphate such as guanosine monophosphate 2-aminothiophosphate (+538.8 m/z), suggesting nucleotidyl transfer in addition to at least one thiophosphoryl transfer. Consistent with this interpretation, the substrate strand became radiolabeled when the ribozyme:substrate complex was incubated with [α-32P]GTP, and radiolabel incorporation was inhibited by excess unlabeled GTP (Figure 4B, lanes 3–9, and Supplementary Figure S7A, lanes 3–9). Thus, the alpha phosphate of [α-32P]GTP is incorporated into the product by a mechanism that competes with unlabeled GTP. Oligonucleotide phosphorylation normally increases electrophoretic mobility due to the additional negative charge, but the product formed by ribozyme K28min migrates more slowly than the unmodified substrate strand in denaturing non-APM gels (Figure 4B, first two lanes), again indicating that the ribozyme catalyzes additional modifications besides thiophosphorylation.
Figure 4.
Identification of nucleotidyl adduct. (A) MALDI-TOF mass spectrometry of input substrate d3.1 (top left), 5΄-thiophosphorylated d3.1 that was generated by treatment with ATPγS and PNK (bottom left), ribozyme-catalyzed product that was formed by treatment of d3.1 with K28min and GTPγS (top right), and acid-decomposition of the ribozyme-catalyzed product (bottom right). The M–H peaks produced by each treatment are indicated: blue triangles, 6784.28 m/z (expected 6782.17 a.m.u for input d3.1 substrate); magenta triangles, 6880.38 m/z (expected 6878.11 a.m.u for mono-thiophosphorylated d3.1); green triangles, 7322.00 m/z (expected 7319.10 a.m.u for adduct shown in panel F top); red triangles, 7225.3 m/z (expected 7223.16 a.m.u for adduct shown in panel F bottom). (B) Incorporation of α-32P label into unlabeled acceptor strand. Controls in first two lanes show 5΄-32P-labeled d3.1 substrate alone (NR) or incubated with ribozyme K28min and GTPγS (sP). For remaining lanes, non-radiolabeled d3.1 substrate was incubated with ribozyme K28min and [α-32P]GTP for 5 min before adding the indicated amount of unlabeled GTP. Reactions were performed at 10°C for 18 h. (C) Thiophosphorylation reactions containing the indicated combinations of ribozyme, substrate and donor were treated with sodium periodate, followed by amination with Cy3 hydrazide dye. For the sample in Lane 4, the product of the ribozyme-catalyzed reaction was purified from an APM gel prior to the dye labeling reaction. Gel was scanned for Cy3 fluorescence (Ex. 532 nm Em. 570 nm). Phosphorimages are of 20% denaturing PAGE.
To determine the state of the ribose moiety, we subjected the product to periodate oxidation, which converts the 2΄-3΄ vicinal diols of a terminal ribose into reactive aldehydes that can then be labeled with a hydrazide dye. As expected, the ribozyme itself appears as a strong, fluorescently-labeled band, and no additional bands were observed when the DNA acceptor substrate was added to the ribozyme prior to this reaction, since DNA is not susceptible to periodate oxidation. In contrast, a new dye-labeled band appeared when the acceptor strand and ribozyme were incubated with GTPγS donor prior to periodate oxidation and dye labeling (Figure 4C). When these treatments were performed using 32P-labeled acceptor DNA strand, the radiolabel and fluorescence signals co-migrated (Supplementary Figure S7B–D). These experiments establish that the DNA acceptor strand acquires a ribonucleotide during the ribozyme-catalyzed reaction, and that both the 2΄OH and 3΄OH remain available for subsequent reactions, consistent with nucleotidyl transfer.
Nature of the product
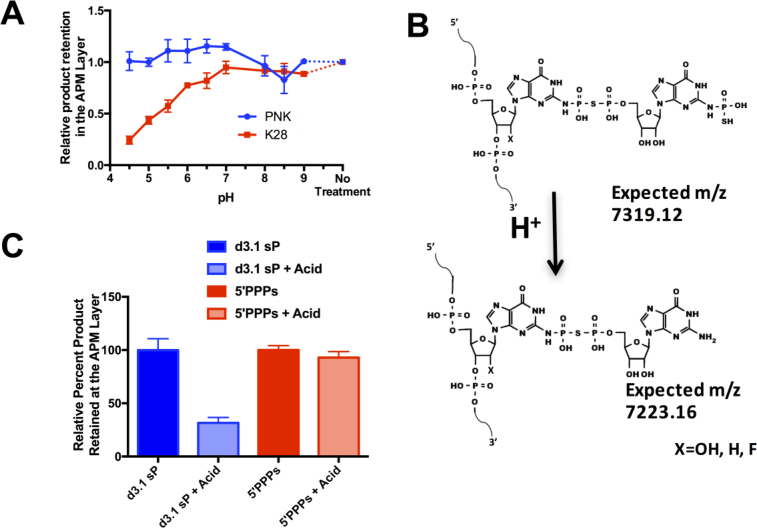
Product decomposition and donor analog reactivity were analyzed in two series of experiments to shed further light on the nature of the adduct and to rule out alternative models. The first series measured product stability as a function of pH. It is known that N-phosphate esters can be more acid labile than O-phosphate esters, often hindering analysis of proteins that carry phosphoarginine, phospholysine (28) or phosphohistidine (29). We reasoned that the product of ribozyme K28min could exhibit similar acid sensitivity. Indeed, the product formed by ribozyme K28min was stable at neutral and elevated pH but unstable under acidic conditions, as measured by loss of APM retention. In contrast, genuine 5΄-mono-thiophosphorylated product formed by polynucleotide kinase was stable at all pH values tested (Figure 5A), as was the 2΄OH thiophosphorylated product of an unrelated ribozyme K37 (1) (Supplementary Figure S8A). Interestingly, acid-treated K28min product was still upshifted relative to the substrate band on denaturing non-APM gels (Supplementary Figure S8B). Acid treatment induced a mass loss of ∼98 m/z (Figure 4A, bottom right), suggesting loss of one thiophosphoryl group but not the GMP portion of the adduct or the other thiophosphoryl group, consistent with the observed electrophoretic mobility. Based on these data, we speculated that the ribozyme may participate in a multi-step reaction to yield an unusual adduct as shown in Figure 5B.
Figure 5.
Product formed by ribozyme K28min is acid labile. (A) Thiophosphorylated product formed either by ribozyme K28min and GTPγS or by T4 polynucleotide kinase and ATPγS was purified from an APM gel, then incubated at the indicated pH for 5 h at 32°C and separated on an APM gel to determine the amount of thiophosphorylated product that remained after treatment at varying pH. (B) Plausible product formed by ribozyme K28min (top) and by its acid-induced dethiophosphorylation (bottom). (C) The product of ribozyme K28min reaction (d3.1 sP) and an RNA transcript carrying a 5΄ γ- thiotriphosphate (5΄PPPs) were each incubated at pH 4.0 or in water, then separated on a denaturing APM gel to determine the amount of thiophosphorylated product remaining after mild acid treatment.
When the reaction was carried out using 100 μM ribozyme:acceptor complex and 2-fold excess (200 μM) GTPγS donor, product peaks accumulated in the MALDI-TOF spectra at both 7321.12 m/z (corresponding to the complete product in Figure 5B) and 7225.30 m/z (corresponding to the dethiophosphorylated product in Figure 5B). The rates of accumulation of materials at each mass were indistinguishable (Supplementary Figure S9), and there was no evidence of one product chasing into the other. This may indicate that both products accumulate simultaneously or in rapid succession, and that a portion of the material may be unable to carry the reaction to completion, or it could reflect partial decomposition during sample workup for MALDI-TOF analysis. No new MALDI-TOF peaks appear that might represent additional intermediates; minor peaks in these spectra were also present before incubation with GTPγS and potentially represent RNA degradation fragments.
The second series of experiments evaluated the alternative possibility that the ribozyme promotes Cu2+-induced formation of 8-oxo-GTPγS, analogous to the Cu2+-mediated oxidation of guanosine that is observed in solution (30). In principle, the C-8 of 8-oxo-GTPγS could condense with the exocyclic N-2 amine of dG2 in the acceptor strand with loss of water to yield a product with a calculated mass of 7319.1 a.m.u., which is only about 3 a.m.u. short of the mass observed in Figure 4A. Such a product would retain an intact ribose ring and would carry an intact γ-thio-triphosphate (PPPs). We therefore generated genuine 5΄-PPPs-RNA transcript and compared its stability to that of the product of ribozyme K28min. Unlike the K28min product, 5΄-PPPs-RNA was stable in acid (Figure 5C). Conversely, treatment with calf intestinal phosphatase rapidly removed thiophosphate from 5΄-PPPs-RNA while having minimal impact on the K28min product (Supplementary Figure S10A-C). Furthermore, K28 was able to generate some product in elevated Mg2+ even when reactions were performed in the presence of reducing agents and in the absence of Cu2+ ions, as analyzed both by APM retention (Supplementary Figure S10D) and by upshift on a non-APM denaturing polyacrylamide gel (Supplementary Figure S10E). These results argue against an alternative reaction involving Cu+2 induced formation of an 8-oxo-G intermediate. Instead, all of the results above are consistent with formation of the unusual nucleobase-modified adduct shown in Figure 5B.
DISCUSSION
Here, we describe an unusual reaction catalyzed by ribozyme K28min involving transfer of two thiophosphoryl groups and a GMP nucleotide onto N-2 of an internal guanosine. This product is indicated by multiple lines of evidence, including the masses of the ribozyme reaction products and their subsequent decomposition products, electrophoretic mobility shifts on denaturing APM and non-APM polyacrylamide gels, 32P label tracing that places both alpha and gamma phosphates of the GTP donor in the final product, phosphatase insensitivity and acid lability of the products of the ribozyme reaction, and chemical reactivity of vicinal diols that indicates an intact ribose. The N-2 of guanosine is known to participate in other chemistries, such as reductive amination by a deoxyribozyme (31) and nucleophilic attack on ribose aldehydes at abasic sites to form interstrand crosslinks (32). N-2 of guanosine also acts as a nucleophile during biosynthesis of modified nucleotides found in tRNA and rRNA such as N2-methyl guanosine (m2G) (33). Other guanosine reactivities are incompatible with the data presented here for ribozyme K28min. For example, in Group I self-splicing introns (34), a guanosine 3΄OH serves as nucleophile to insert into the RNA strand and displace the 5΄ exon. However, a Group I-like reaction would preclude the observed periodate oxidation of the product and would reduce the mass of the product upon loss of the leaving group. Similarly, although RNA-catalyzed alkylation of guanosine N-7 has been reported (35), N-7 reactivity cannot be a requirement for the K28min reaction, as acceptor substrates carrying 7-deaza substitution at the acceptor site remain able to react. Instead, the guanosine N-7 and O-6 functional groups may help to stabilize the loosely-organized active site. Finally, an alternative model involving an 8-oxo-GTPγS intermediate was ruled out by differences in phosphatase and acid sensitivities between genuine 5΄-PPPs-RNA and the product of the ribozyme reaction and by continued low-level product formation under reducing conditions and in the absence of Cu2+.
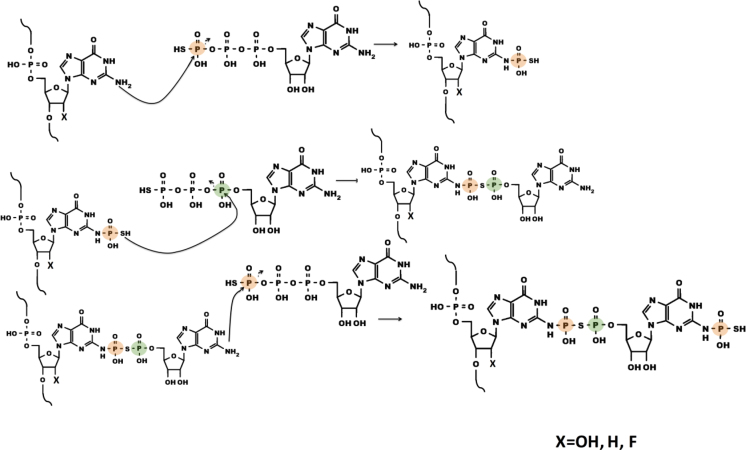
Although the precise mechanism of this ribozyme remains to be determined, the data presented here are consistent with the formation of a larger ribonucleotide adduct on the nucleobase of the substrate oligonucleotide. The product formed by the ribozyme could potentially arise from a three-step mechanism (Figure 6). In the first step, the exocyclic amine of dG2 in the substrate strand attacks the gamma thiophosphate of GTPγS, displacing GDP as leaving group. That the adduct initiates at this position in the acceptor strand is supported by product formation for dG2→(2AP) and dG2→(7deazaG) substitutions and by the lack of product formation for dG2I. In the second step, the thiophosphate attacks the α-phosphorus of GDP or another GTPγS donor, displacing inorganic phosphate or thio-pyrophosphate as a leaving group and generating a GDP(β)S adduct. The predicted mass of this species exactly matches that observed for the acid decomposition product and for the minor product from kinetic studies at low donor concentration. Lack of APM retention for the acid decomposition product suggests that the sulfur from the first step is inaccessible after mild acid treatment, which would be the case if it is the nucleophile for the second step of the ribozyme reaction. The presence of intact ribose hydroxyls is indicated by the susceptibility of the product of the ribozyme reaction to periodate oxidation and subsequent labeling with dye. In the third step, the exocyclic amine of the GDP(β)S adduct attacks the gamma thiophosphate of another GTPγS, again displacing GDP as the leaving group. No additional products of higher masses are evident by mass spectrometry; this could potentially be due to the disruption of the active site after the final step. No intermediates were observed, which could indicate that the first step of the reaction is rate limiting, with rapid subsequent steps. As noted above, this second thiophosphoryl group is proposed to be the target of acid decomposition. The ribozyme's lack of reactivity with, or recognition of, ITPγS precluded the use of this analog in assessment of reaction intermediates (Supplementary Figure S11). Only a small handful of in vitro selected ribozymes have been previously observed to carry out multi-step reactions, such as the addition of phenylalanine first onto a ribozyme 3΄-terminal 2΄OH and then further Phe additions onto a growing poly-Phe chain (36). The current work provides the first indication of multi-step, sequential reactions for a ribozyme that appears to utilize both nucleotidyl and (thio)phosphoryl transfers.
Figure 6.
Speculative three-step reaction mechanism. Details in text. Orange and green circles track the γ-P and the α-P of donor GTPγS, respectively.
Phosphoryl transfer onto nucleobases occurs in several biological settings, including adenosine biosynthesis, during which O-6 phosphorylation of inosine monophosphate activates the oxygen for displacement by the α-NH2 of aspartate (37). Similarly, the O-4 of uracil is phosphorylated during the biosynthesis of cytosine (38). N-6 of adenosine has the potential to act as a nucleophile during the biosynthesis of N-6-threonylcarbamoyladenosine (t6A) modified tRNA (39). Chemical synthesis of N-2 phosphorylated guanosine has been reported (40), and although it has not been observed as a biological metabolite, N-phosphorylation of other small metabolites is common in biology; for example, phosphohistidine plays critical roles in signal processing, while phosphocreatine (41), phosphoarginine and phospholysine (28) are all utilized as energy sources. Thus, the unique and unprecedented reactions catalyzed by ribozyme K28min are analogous to classes of reactions that are critical for biology. Furthermore, they raise the possibility that similar ribozymes could support metabolic reactions and further diversify nucleobase chemical reactivity, whether in extant, engineered or incipient life forms.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elisa Biondi, Mark Ditzler, Khalid Alam, and Kent Gates for feedback on the manuscript. We would also like to thank Steven Benner, Frank Schmidt, Peter Tipton and members of the Burke lab for helpful discussions.
Author contributions: R.R.P., D.H.B. and R.K. conceived experiments and designed research. R.R.P., P.D-M.N, M.P.L, M.K.C. and J.V.G. performed experiments. R.R.P. and D.H.B. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NASA Earth Science Space Fellowship [NNX13AP86H to R.R.P.]; NASA Exobiology [NNX12AD66G to D.H.B.]. Funding for open access charge: NASA [NNX12AD66G to D.H.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Biondi E., Nickens D., Warren S., Saran D., Burke D.H.. Convergent donor and acceptor substrate utilization among kinase ribozymes. Nucleic Acids Res. 2010; 36:6785–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorsch J.R., Szostak J.W.. In vitro evolution of new ribozymes with polynucleotide kinase activity. Nature. 1994; 371:31–36. [DOI] [PubMed] [Google Scholar]
- 3.Seelig B., Jäschke A.. A small catalytic RNA motif with Diels-Alderase activity. Chem. Biol. 1999; 6:167–176. [DOI] [PubMed] [Google Scholar]
- 4.Joyce G.F. The antiquity of RNA-based evolution. Nature. 2002; 418:212–221. [DOI] [PubMed] [Google Scholar]
- 5.Chen X., Li N., Ellington A.. Ribozyme catalysis of metabolism in the RNA world. Chem. Biodivers. 2007; 4:633–655. [DOI] [PubMed] [Google Scholar]
- 6.Burke D.H. de Pauplana LR. Genetic Code and the Origin of Life. 2004; Landes Bioscience. [Google Scholar]
- 7.Lorsch J.R., Szostak J.W.. Kinetic and thermodynamic characterization of the reaction catalyzed by a polynucleotide kinase ribozyme. Biochemistry. 1995; 34:15315–15327. [DOI] [PubMed] [Google Scholar]
- 8.Saran D., Nickens D., Burke D.. A trans acting ribozyme that phosphorylates exogenous RNA. Biochemistry. 2005; 44:15007–15016. [DOI] [PubMed] [Google Scholar]
- 9.Saran D., Held D., Burke D.. Multiple-turnover thio-ATP hydrolase and phospho-enzyme intermediate formation activities catalyzed by an RNA enzyme. Nucleic Acids Res. 2006; 34:3201–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biondi E., Nickens D.G., Warren S., Saran D., Burke D.H.. Convergent donor and acceptor substrate utilization among kinase ribozymes. Nucleic Acids Res. 2010; 38:6785–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis E., Bartel D.. New catalytic structures from an existing ribozyme. Nat. Struct. Mol. Biol. 2005; 12:994–1000. [DOI] [PubMed] [Google Scholar]
- 12.Li Y., Breaker R.R.. Phosphorylating DNA with DNA. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:2746–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Billen L.P., Li Y.. Sequence diversity, metal specificity, and catalytic proficiency of metal-dependent phosphorylating DNA enzymes. Chem. Biol. 2002; 9:507–517. [DOI] [PubMed] [Google Scholar]
- 14.Schlosser K., Lam J., Li Y.. A genotype-to-phenotype map of in vitro selected RNA-cleaving DNAzymes: implications for accessing the target phenotype. Nucleic Acids Res. 2009; 37:3545–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camden A.J., Walsh S.M., Suk S.H., Silverman S.K.. DNA oligonucleotide 3΄-phosphorylation by a DNA enzyme. Biochemistry. 2016; 55:2671–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorsch J.R., Bartel D., Szostak J.W.. Reverse transcriptase reads through a 2΄-5΄ linkage and a 2΄-thiophosphate in a template. Nucleic Acids Res. 1995; 23:2811–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh S.M., Sachdeva A., Silverman S.K.. DNA catalysts with tyrosine kinase activity. J. Am. Chem. Soc. 2013; 135:14928–14931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelinas A.D., Davies D.R., Edwards T.E., Rohloff J.C., Carter J.D., Zhang C., Gupta S., Ishikawa Y., Hirota M., Nakaishi Y. et al. . Crystal structure of interleukin-6 in complex with a modified nucleic acid ligand. J. Biol. Chem. 2014; 289:8720–8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta S., Hirota M., Waugh S.M., Murakami I., Suzuki T., Muraguchi M., Shibamori M., Ishikawa Y., Jarvis T.C., Carter J.D. et al. . Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J. Biol. Chem. 2014; 289:8706–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaught J.D., Bock C., Carter J.D., Fitzwater T., Otis M., Schneider D., Rolando J., Waugh S.M., Wilcox S.K., Eaton B.E.. Expanding the Chemistry of DNA for In Vitro Selection. J. Am. Chem. Soc. 2009; 132:4141–4151. [DOI] [PubMed] [Google Scholar]
- 21.Fabrega C., Hausmann S., Shen V., Shuman S., Lima C.D.. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell. 2004; 13:77–89. [DOI] [PubMed] [Google Scholar]
- 22.Poudyal R.R., Callaway M.K., Benslimane M., Lokugamage M.P., Staller S., Burke D.H.. Selective Inactivation of functional RNAs by ribozyme-catalyzed covalent modification. ACS Synth. Biol. 2016; doi:10.1021/acssynbio.6b00222. [DOI] [PubMed] [Google Scholar]
- 23.Biondi E., Burke D.H.. Separating and analyzing sulfur-containing RNAs with organomercury gels. Methods Mol. Biol. 2012; 883:111–120. [DOI] [PubMed] [Google Scholar]
- 24.Koomen J.M., Russel W.K., Hettick J.M., Russel D.H.. Improvement of resolution, mass accuracy, and reproducibility in reflected mode DE-MALDI-TOF analysis of DNA using fast evaporation-overlayer sample preparations. Anal. Chem. 2000; 72:3860–3866. [DOI] [PubMed] [Google Scholar]
- 25.Biondi E., Poudyal R.R., Forgy J., Sawyer A., Maxwell A., Burke D.H.. Lewis acid catalysis of phosphoryl transfer from a copper(II)-NTP complex in a kinase ribozyme. Nucleic Acids Res. 2013; 41:3327–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biondi E., Maxwell A., Burke D.H.. A small ribozyme with dual-site kinase activity. Nucleic Acids Res. 2012; 40:7528–7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pallan P.S., Greene E.M., Jicman P.A., Pandey R.K., Manoharan M., Rozners E., Egli M.. Unexpected origins of the enhanced pairing affinity of 2΄-fluoro-modified RNA. Nucleic Acids Res. 2011; 39:3482–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basant P.G., Attwood P.V., Pigott M.J.. Focus on phosphoarginine and phospholysine. Curr. Protein Pept. Sci. 2009; 10:536–550. [DOI] [PubMed] [Google Scholar]
- 29.Kee J.M., Muir T.W.. Chasing phosphohistidine, an elusive sibling in the phosphoamino acid family. ACS Chem. Biol. 2012; 7:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd D., Phillips D.. Oxidative DNA damage mediated by copper(II), iron(II) and nickel(II)Fenton reactions: evidence for site-specific mechanisms in the formation of double-strand breaks, 8-hydroxydeoxyguanosine and putative intrastrand cross-links. Mut. Res. 1999; 424:23–36. [DOI] [PubMed] [Google Scholar]
- 31.Wong O.Y., Mulcrone A.E., Silverman S.K.. DNA-catalyzed reductive amination. Angew. Chem. 2011; 50:11679–11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson K.M., Price N.E., Wang J., Fekry M.I., Dutta S., Seiner D.R., Wang Y., Gates K.S.. On the formation and properties of interstrand DNA-DNA cross-links forged by reaction of an abasic site with the opposing guanine residue of 5΄-CAp sequences in duplex DNA. J. Am. Chem. Soc. 2013; 135:1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ihsanawati, Nishimoto M., Higashijima K., Shirouzu M., Grosjean H., Bessho Y., Yokoyama S.. Crystal structure of tRNA N2, N2-guanosine dimethyltransferase Trm1 from Pyrococcus horikoshii. J. Mol. Biol. 2008; 383:871–884. [DOI] [PubMed] [Google Scholar]
- 34.Zaug A.J., Cech T.R.. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA. 1995; 1:362–374. [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson C., Szostak J.W.. In Vitro evolution of a self-alkylating ribozyme. Nature. 1995; 374:777–782. [DOI] [PubMed] [Google Scholar]
- 36.Turk R.M., Chumachenko N.V., Yarus M.. Multiple translational products from a five-nucleotide ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4585–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bass M.B., Fromm H.J., Rudolph F.B.. The mechanism of the adenylosuccinate synthetase reaction as studied by positional isotope exchange*. J. Biol. Chem. 1984; 259:12330–12333. [PubMed] [Google Scholar]
- 38.Levitzki A., Koshland D.E.J.. Cytidine triphosphate synthetase. Covalent intermediates and mechanisms of action. Biochemistry. 1971; 10:3365–3371. [DOI] [PubMed] [Google Scholar]
- 39.Perrochia L., Crozat E., Hecker A., Zhang W., Bareille J., Collinet B., van Tilbeurgh H., Forterre P., Basta T.. In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya. Nucleic Acids Res. 2013; 41:1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wada T., Moriguchi T., Sekine M.. Synthesis and properties of N-phosphorylated eibonucleosides. J. Am. Chem. Soc. 1994; 116:9901–9911. [Google Scholar]
- 41.McLeish M.J., Kenyon G.L.. Relating structure to mechanism in creatine kinase. Crit. Rev. Biochem. Mol. Biol. 2005; 40:1–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.