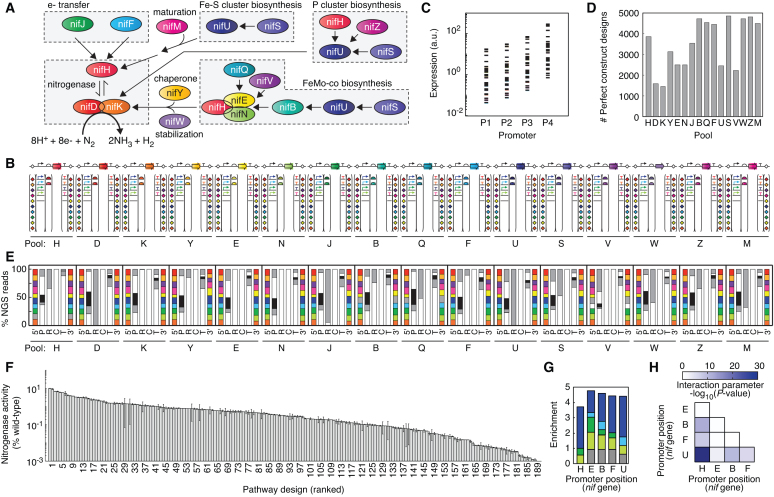
Figure 3.
Algorithm-guided exploration of genetic design space for a 16-gene nitrogen fixation pathway. (A) An overview of the K. oxytoca pathway (65). (B) Pool designs for composite parts with one nif gene in each pool. Parts were permuted as shown with variants of promoters (5), RBSs (2), terminators (5) and assembly linker (10 × 10) parts in each nif pool. Across the 16 pools, there are 96 part positions in total. Pool names are abbreviated by last letter of gene name. (C) Expression range of promoter-RBS combinations. Expected expression of promoter and RBS part combinations for the design space in the pools. Expression was estimated as the product of promoter strength (REU units) (65) and RBS strength (67,68). (D) Number of sequence-perfect composite part designs in nif pools. A cutoff of at least 1 sequence-perfect construct for each design was applied. (E) Representation of parts in the sequence-perfect constructs (% NGS reads that perfectly match a composite part design) at each position in the 16 pools. Approximately 4 million NGS reads per pool were sequenced and analyzed. Part type abbreviations: 5΄ linker (5΄), promoter (P), RBS (R), CDS (C), terminator (T), 3΄ linker (3΄). Linker positions are colored according to the order listed in Supplementary Data and parts variants at each position (bottom to top) follow the order listed. (F) Assayed activity for a 192-member algorithm-designed library built from the pools. Acetylene substrate was supplied, and ethylene produced was measured by GC analysis. Nitrogenase activity (% wild-type) is calculated using assayed K. oxytoca ethylene production. Error bars show deviation of duplicate assays for each pathway design. (G) Part enrichment for the top 20 pathway designs as compared to the whole 192-member library. Enrichment for each promoter position was calculated based on its frequency (freqTop20/freqLibrary). The nif genes were arranged in the order shown in panel B. The promoter variants include PT7.4 (purple), PT7.3 (blue), PT7.2 (green) and PT7.1 (yellow) and P0 (grey) with sequences listed in Supplementary Data. (H) Significance of pairwise interaction parameters in the fitted response surface model for nitrogenase activity (Methods).