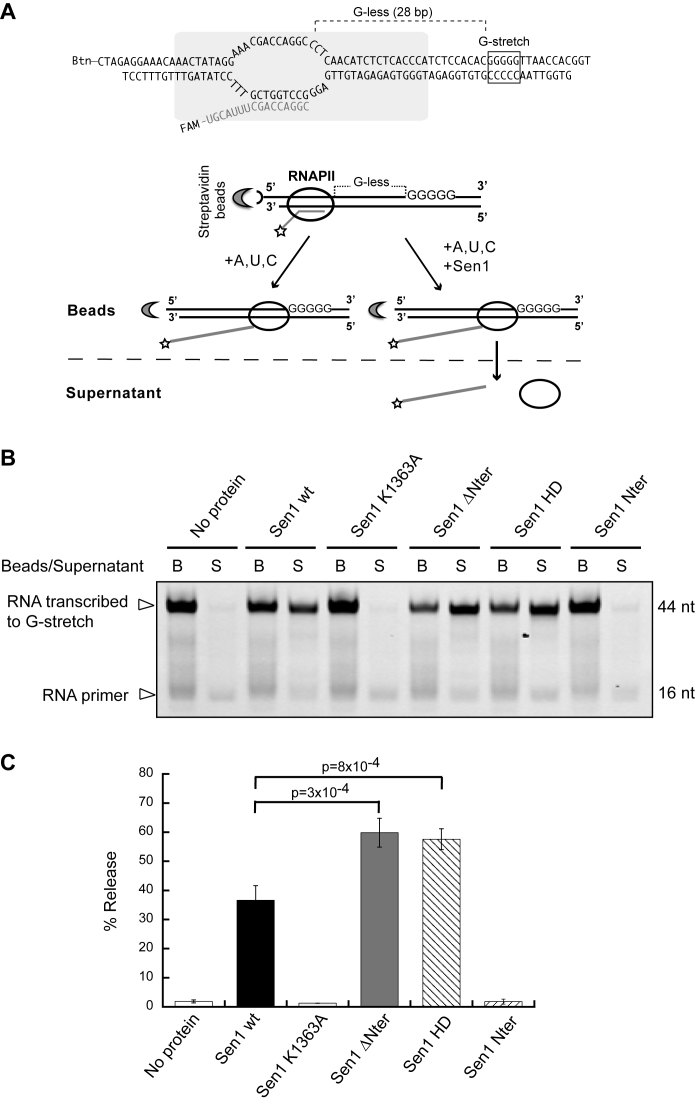
Figure 2.
Analysis of the capability of Sen1 variants to terminate transcription in vitro. (A) Scheme of an in vitro transcription termination assay. A schematic of a ternary EC based on previous biochemical and structural analyses (47,48) is shown on the top. Promoter-independent assembly of ECs is performed using a 9 nt RNA:DNA hybrid that occupies the RNAPII catalytic center. Ternary ECs are attached to streptavidin beads via the 5΄ biotin of the non-template strand allowing subsequent separation of bead-associated (B) and supernatant (S) fractions. The RNA is fluorescently labeled with FAM at the 5΄-end. The transcription template contains a G-less cassette followed by a G-stretch in the non-template strand. After adding an ATP, UTP, CTP mix, the RNAPII transcribes until it encounters the G-rich sequence. Sen1 provokes dissociation of ECs paused at the G-rich stretch and therefore the release of RNAPII and associated transcripts to the supernatant. (B) Denaturing PAGE analysis of RNAs from a representative IVTer assay in the absence and in the presence of Sen1 proteins. (C) Quantification of the fraction of transcripts released from ECs stalled at the G-stretch as a measure of the termination efficiency. Values represent the average and standard deviation of three independent experiments. The p-value associated with a t-test (p) is indicated.