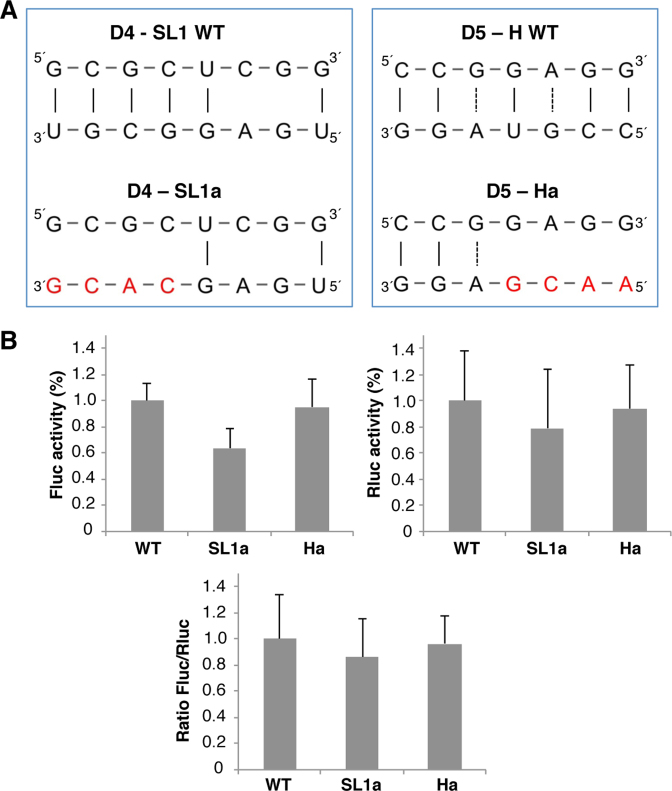
Figure 9.
Functional analysis of the 3΄UTR sequences predicted to base pair with the IRES element. (A) Diagram of the base pairs disrupted between the domain 4 or domain 5 of the IRES and the 3΄UTR in constructs SL1a or Ha. Red letters depict nucleotide substitutions of the 3΄UTR sequence. (B) Translation efficiency of the SL1a and Ha constructs. The Firefly luciferase (Fluc) activity/μg of protein and Renilla luciferase (Rluc) activity/μg of protein was made relative to the values obtained for the wild type construct performed in parallel. Relative IRES activity (mean ± SD) was determined in transfected BHK-21 cells as the Fluc/Rluc ratio expressed from monocistronic constructs, then normalized to the activity observed for the wild type IRES (set to 1). Each experiment was performed in triplicate and repeated three times.