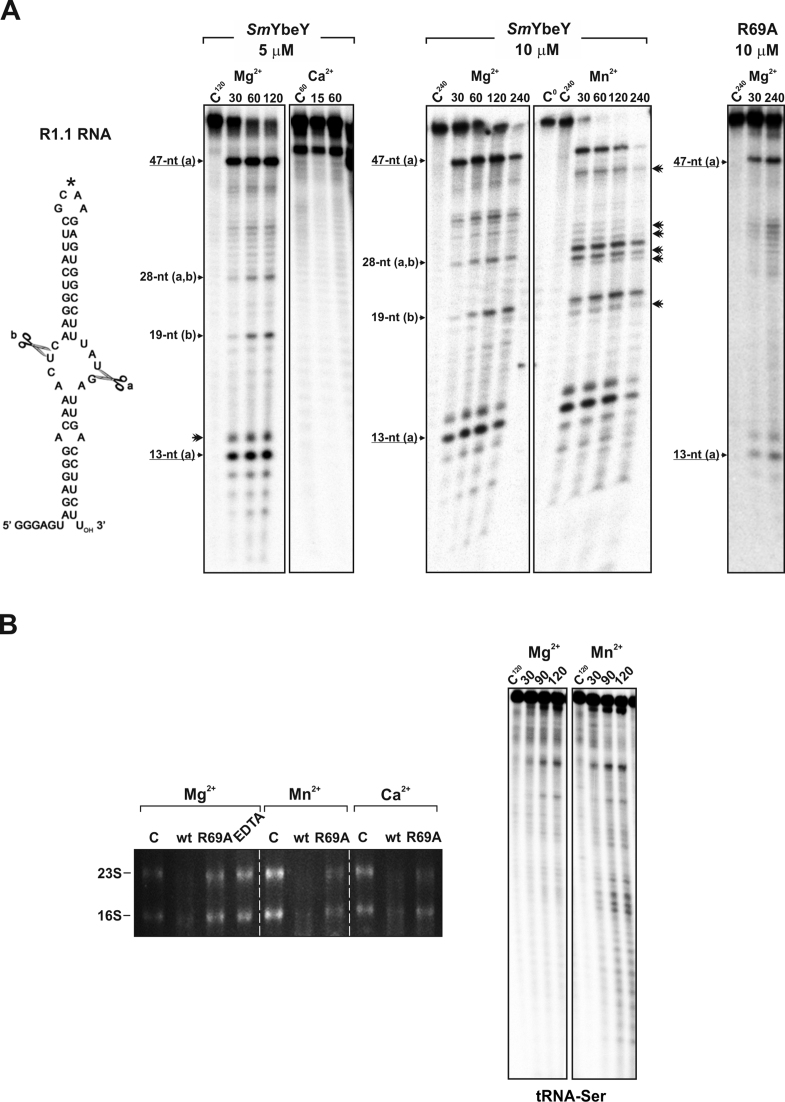
Figure 4.
Activity of SmYbeY on structured RNA substrates. (A) Reactivity patterns of SmYbeY and SmYbeY-R69A on the R1.1 RNA (1.12 pmol/μl), the canonical substrate for RNase III. Sequence and secondary structure of the substrate is shown to the left. Enzyme concentrations, metal co-factor and reaction times are indicated on top of the panels. Reactions were analyzed on 7 M urea/15% polyacrylamide gels. Known cleavage sites (A and B) of RNase III on the same substrate are indicated. The asterisk (*) indicates an alternative minor cleavage site within R1.1. Additional minor reactions products when Mn2+ was used as co-factor are indicated by double arrowheads. C, control reactions. (B) Left panel, activity of SmYbeY and SmYbeY-R69A on rRNA (500 ng). Enzymes (20 μM) were incubated for 2 h at 37°C in the presence of EDTA (50 mM) and/or the co-factors indicated on top. RNA was analyzed on a 1.5% agarose gel. The positions of the 23S and 16S rRNA are indicated. C, control reactions. Right panels, activity of SmYbeY (10 μM) on tRNA-Ser. Metal co-factors used in the assays and the time-course of the reactions are indicated on top of the panels. The reaction products were separated on 7 M urea/10% polyacrylamide gels. C, control reactions.