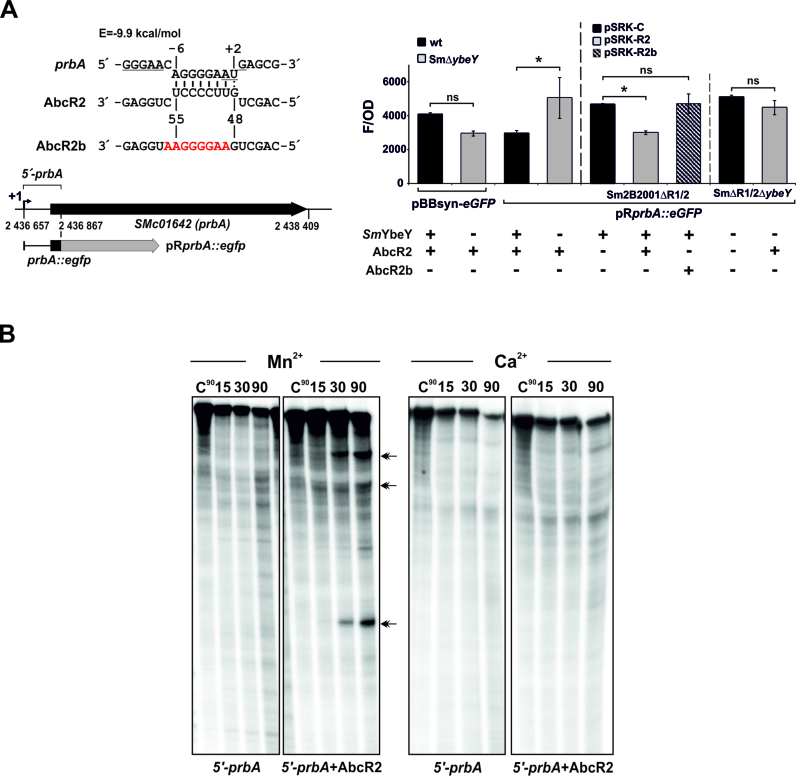
Figure 9.
SmYbeY is required for the AbcR2-mediated silencing of the prbA mRNA. (A) Lack of SmYbeY abrogates AbcR2-dependent post-transcriptional repression of prbA in vivo. The AbcR2-prbA antisense interaction as predicted by IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) is shown on top. The Shine–Dalgarno sequence and AUG start codon of prbA are underlined. Numbers indicate positions relative to the prbA start codon and AbcR2 transcription start site. The predicted minimum hybridization energy (E) of the duplex is indicated. Nucleotide substitutions in the AbcR2b mutant variant disrupting base-pairing with prbA are colored in red. The schematics below show the prbA genomic region and the reporter fusion used in the in vivo assays. Numbers stand for coordinates in the genome of the reference strain Rm1021. The transcription start site of prbA is indicated (+1). Histogram; fluorescence of the Sm2B3001 strain and its SmΔybeY mutant derivative transformed with the control plasmid pBBsyn-eGFP or the reporter pRprbA::eGFP (left part) and fluorescence of the double Sm2B2001ΔR1/2 and triple SmΔR1/2ΔybeY mutants co-transformed with pRprbA::eGFP and pSRK-C (control), pSRK-R2 or pSRK-R2b. Values reported are means and standard deviation of 27 fluorescence measurements normalized to the OD of the culture, i.e. three determinations of three independent exponential cultures of three transconjugants for each reporter strain. The statistical significance level was set at P < 0.05 and indicated with an asterisk. ns, non-significant. SmYbeY, AbcR2 and AbcR2b genotypes of the reporter strains are indicated on the bottom of the histogram. (B) SmYbeY cleaves the AbcR2-prbA duplex in vitro. Reactivity patterns of SmYbeY (5 μM) on an internally labeled 5΄-prbA RNA (0.1 pmol/μl) either alone or in the presence of a molar excess of AbcR2. Double arrow heads indicate major cleavage products. Reactions were incubated in the presence of Mn2+ or Ca2+ as co-factor. Incubation times are indicated on top of the panels. Reactions were analyzed on 7 M urea/15% polyacrylamide gels. C, control reactions.