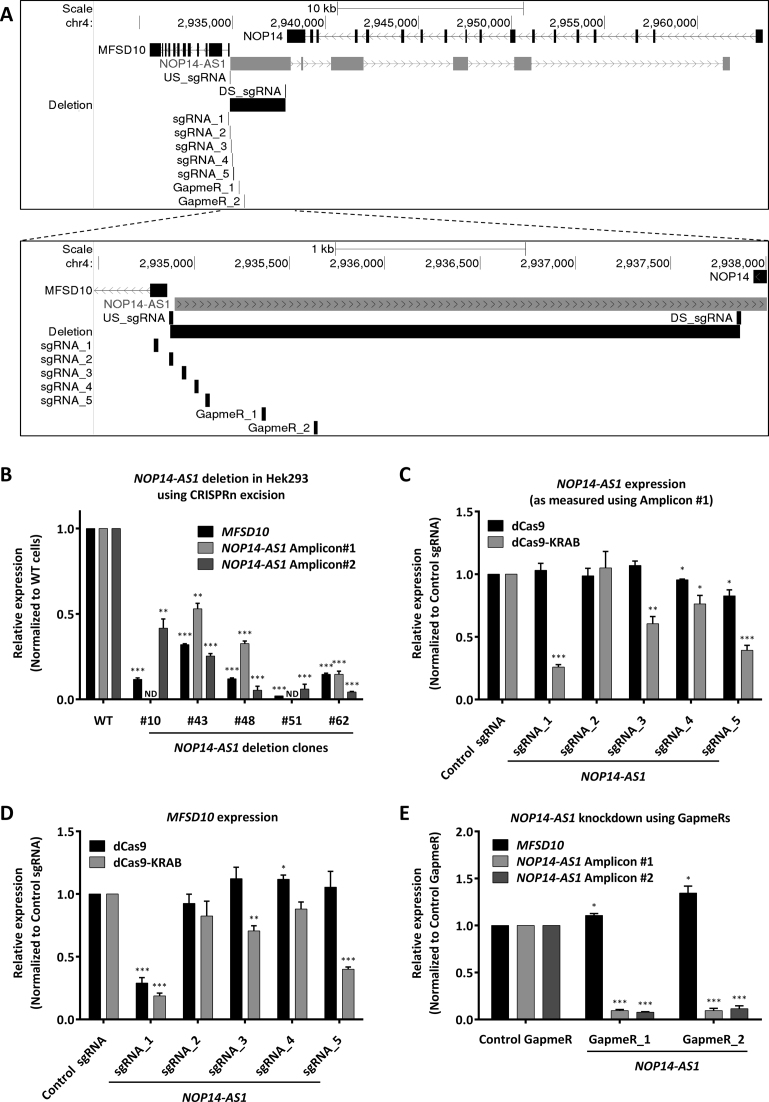
Figure 3.
NOP14-AS1 gene modulation using CRISPRn / CRISPRi and antisense LNA GapmeRs. (A) Schematic representation of the MFSD10 / NOP14-AS1 genomic locus depicting the sgRNAs and ASOs used in this study to target NOP14-AS1. (B) Expression analysis of NOP14-AS1 deletion clones in HEK293 cells. RT-qPCR results for MFSD10 and NOP14-AS1 (amplicon #1 and amplicon #2) normalized to cyclophilin A and Wild Type (WT) cells. ND = not detected. Error bars represent SEM (n = 3). * P < 0.05; ** P < 0.01; *** P < 0.001 compared to WT, unpaired two-sided t-test. (C, D) NCI-H460 cells expressing either dCas9 or dCas9-KRAB were transduced with either Control sgRNA or one of the five indicated sgRNAs targeting NOP14-AS1. RT-qPCR results for NOP14-AS1 amplicon #1 (C) and MFSD10 (D) normalized to cyclophilin A and control sgRNA. Error bars represent SEM (n ≥ 4). *P < 0.05; **P < 0.01; ***P < 0.001 compared to dCas9 / dCas9-KRAB + control sgRNA, unpaired two-sided t-test. (E) NCI-H460 cells were transfected with either a control or two independent antisense LNA GapmeRs against NOP14-AS1. RT-qPCR results for NOP14-AS1 amplicon#1, amplicon#2 and MFSD10 normalized to cyclophilin A and control antisense LNA GapmeR. Error bars represent SEM (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001 compared to control GapmeR, unpaired two-sided t-test.