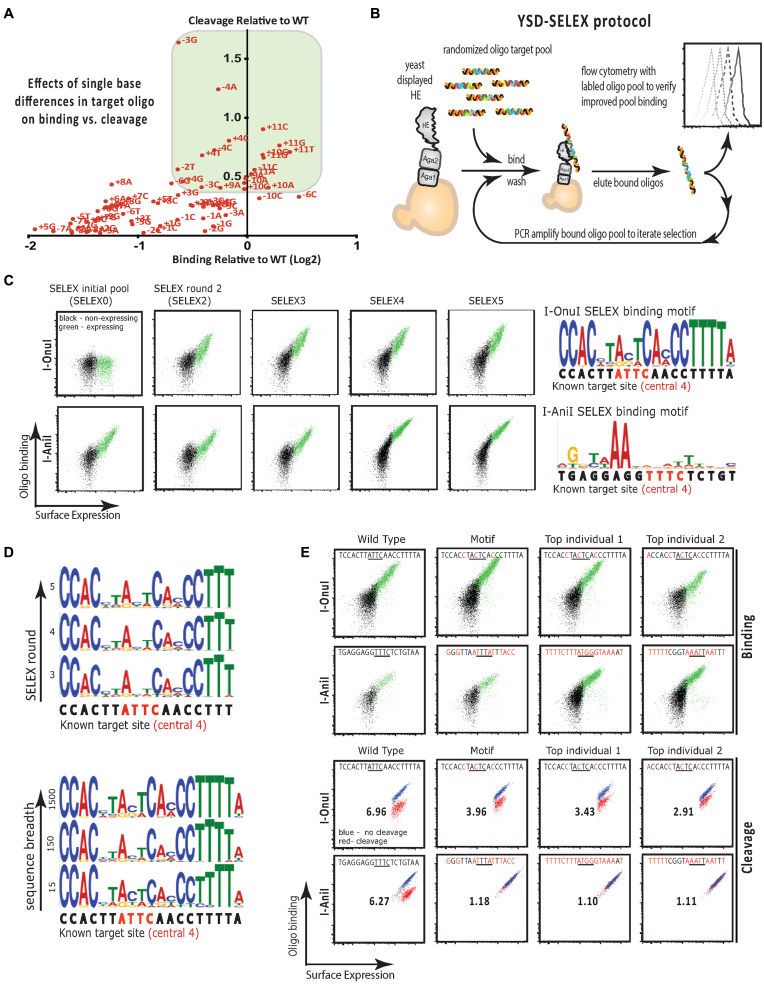
Figure 1.
(A) Plot of binding (Log2(x/WT)) versus cleavage (Ca++/Mg++ normalized to WT) for I-OnuI ‘one-off’ oligonucleotide targets – targets that differ by one base from the native I-OnuI target site at the indicated position. Targets that exhibit high binding (green box) are also typically cleaved efficiently. (B) Schematic of SELEX using yeast surface displayed protein. The homing endonuclease of interest is expressed on the surface of yeast. The yeast is used as a solid support for the protein to facilitate wash steps after binding of randomized oligos. The best-bound sequences are extracted by heating and collecting the supernatant, amplified and subjected to iterative rounds of selection. The initial optimization and later, the success of the experiment are assayed using fluorescently labeled pools and flow cytometry. (C) Flow cytometry plots obtained from staining expressing yeast (green) with labeled target oligonucleotide pools from the indicated SELEX round for I-OnuI and I-AniI. The motif generated by sequencing and analysis of the SELEX5 pool is shown in the far right panel. (D) Representative motif sequence logos for I-OnuI showing that the same profile is obtained by sequence analysis of pools obtained in SELEX oligonucleotide pools 3–5 (top panel), and at very different sequencing breadths (bottom panel). (E) Flow cytometry binding (top) and cleavage analysis (bottom) of WT target oligonucleotides (first column), and targets corresponding to the SELEX motif and the top two most frequent individual sequences obtained from sequencing of the SELEX5 pool. The number in the center of each cleavage plot is the ratio of the mean fluorescence intensity obtained following incubation of the assay with Ca2+ as the divalent ion (non-cleaving conditions) or Mg2+ as the divalent ion (cleaving conditions), and provides a quantitative assessment of the extent of cleavage by the surface expressed enzyme.