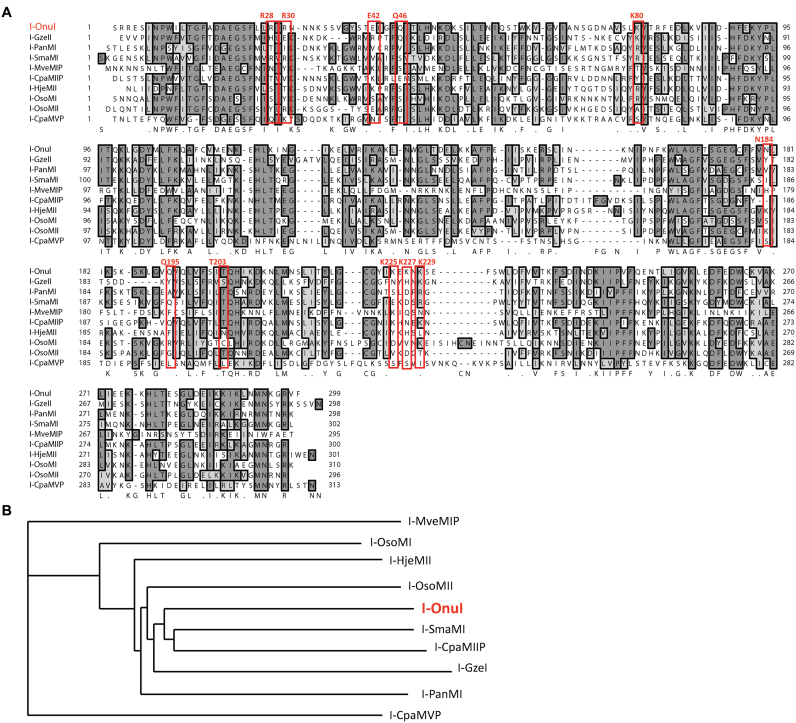
Figure 2.
SELEX homing endonuclease set. (A) Alignment of I-OnuI with benchmark (I-SmaMI, I-PanMI and I-GzeII) and test (I-HjeMII, I-OsoMI, I-OsoMII, I-CpaMIIP, I-CpaMVP and I-MveMIP, LHEs). Residues making direct contact with target site bases in the I-OnuI crystal structure, and the residues that align with them, are boxed in red to emphasize the diverse recognition sequences and mechanisms represented in the group. (B) The phylogram resulting from a ClustalW2 multiple alignment of the protein sequences of the set of LHEs in the SELEX experiments.