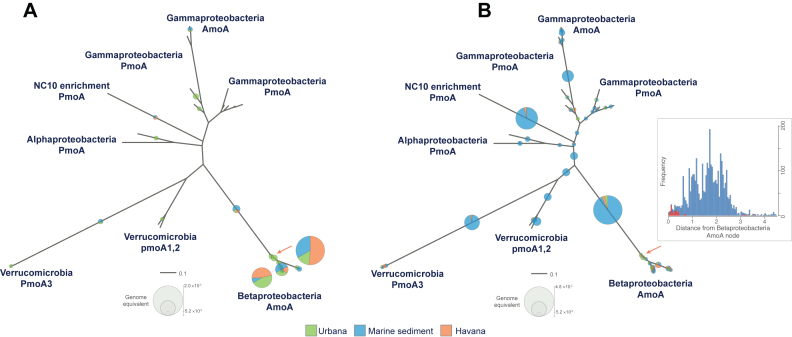
Figure 5.
Placement of amoA reads recovered from terrestrial and marine metagenomes in an AmoA and PmoA phylogenetic tree. A total of 27 bacterial AmoA and PmoA sequences available in the public databases were used to build a reference phylogenetic tree. ROCker bacterial AmoA profiles including (left panel) and not including negative bacterial PmoA references (right panel) were used to identify amoA reads from three metagenomes (see figure key). Reads were placed in the phylogenetic tree using RAxML EPA. The radii of the pie charts represent the abundance for each node (calculated as genome equivalents). Note that most reads in the left tree were placed in the betaproteobacterial AmoA clade. However, the reads in the right tree were placed in more deep-branching nodes of the tree or PmoA clades. The inset shows the distribution of the evolutionary distances of the reads from the (target) betaproteobacterial AmoA node (orange arrow), obtained when a ROCker profile including (red bars) and not including (blue bars) negative references was used.