FIG. 1.
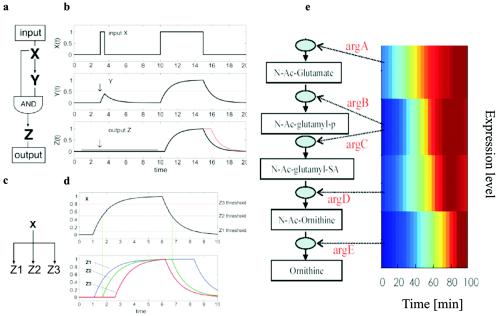
Transcriptional network motifs. Complex transcription networks are made out of simple network motifs (31, 36): patterns that recur throughout the network much more frequently than in randomized networks. Each of the network motif circuits has a defined information processing role. The feed-forward loop circuit (a), which appears in hundreds of systems from bacteria to humans, acts as a persistence detector (27) (b): it filters out short input pulses and responds only to persistent signals. The single-input module (c) can generate temporal expression programs. It allows serial expression of genes by means of differential activation thresholds by the master regulator (d). The single-input module in the arginine biosynthesis system of E. coli (e) was experimentally found to generate a temporal order of genes by means of high-resolution expression measurement employing fluorescent reporter strains (49). Strikingly, the temporal order corresponds to the order of the gene products in the metabolic pathway: the closer the gene to the beginning of the pathway, the earlier its expression. Thus, evolution uses a just in time production strategy, similar to engineering principles of production pipelines. (Figure kindly provided by Uri Alon.)