FIG. 7.
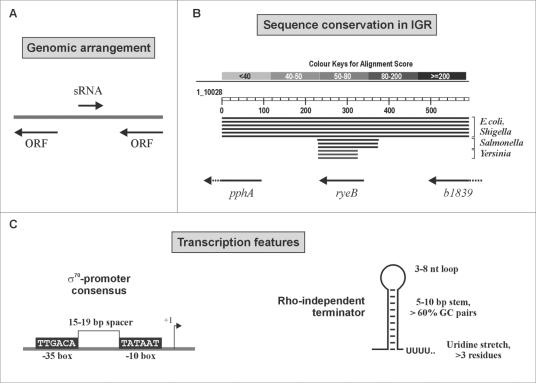
Biocomputational screens have employed individual sRNA features for predictions of these genes in bacteria. The predictive scheme used by Argaman et al. was used to search the “empty” intergenic regions of E. coli for new sRNA genes. (A) sRNA candidates in intergenic regions that were orientated oppositely to both neighboring genes were considered particularly “safe”; that is, they could not correspond to an mRNA leader or trailers. (B) Since many sRNA genes are conserved among closely related bacteria, comparative analysis of intergenic sequences has been used with great success for sRNA identification. Shown is a BLASTN search result for the intergenic region, including 100 bp of adjoining coding sequence, that harbors the sRNA gene ryeB (45). Conservation of the entire region from E. coli MG1655 is limited to the genomes of pathogenic E. coli and Shigella strains. However, the ryeB sequence itself displays high homology values in related enterobacteria such as Salmonella and Yersinia species. (C) Transcription features shared by many E. coli sRNAs that served as input to search for “orphan” promoter and terminator sequences in intergenic regions.