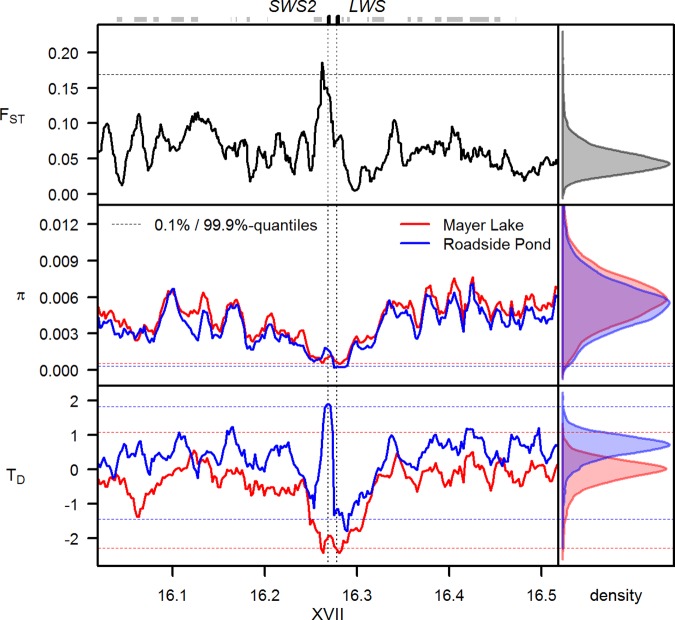
Fig 3. Signature of selective sweep in blackwater-adapted stickleback and “reversed sweep” after transplant to clearwater habitat.
Significantly reduced nucleotide diversity (middle left) and Tajima’s D (TD, bottom left) in the blackwater source population Mayer Lake compared to genome-wide expectations (right) indicate a selective sweep in this population, consistent with the signature seen across the adaptive radiation (cf. Figs 1 and 2). After transplant to a clearwater habitat, however, the alternate haplotype has increased in frequency (Fig 2), leading to one of the strongest differentiation signals in the genome (FST, top left panel) and a significantly positive Tajima’s D. Such a pattern is consistent with a transient phase of a reversed selective sweep. Dashed horizontal lines indicate the 0.1%- and 99.9%-quantiles for each statistic based on their genome-wide distribution in regions with similar recombination rates (right panels); boxes above the figure indicate the position of genes, with black boxes and vertical dashed lines highlighting the position of the two cone opsins SWS2 and LWS. Sliding-window FST, nucleotide diversity and Tajima’s D values and genome-wide distributions of each statistic can be found in S1 Data.