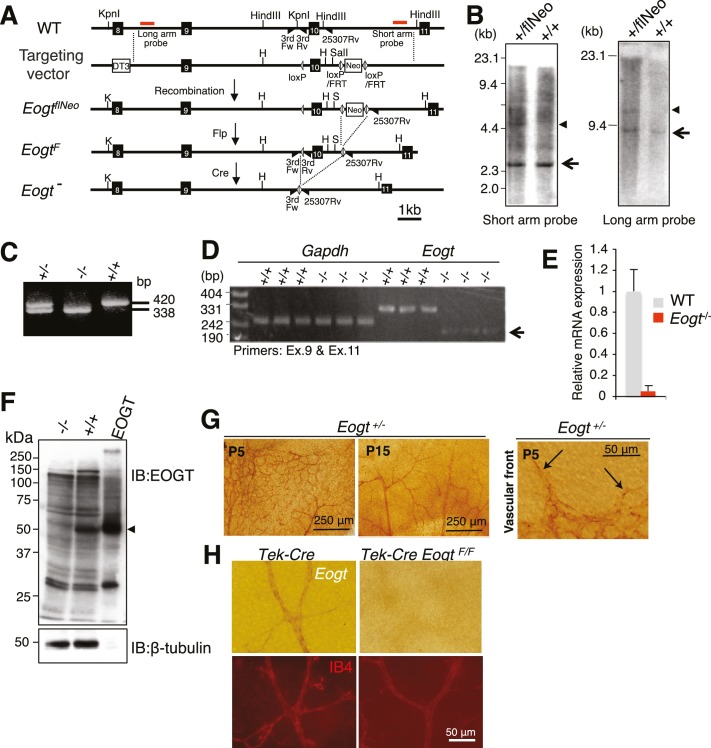
Figure 4. Generation of Eogt-targeted mice.
(A) Schematic drawing of the wild-type mouse Eogt allele (WT), the targeting vector, the floxed allele with the neomycin (Neo)-resistance gene (EogtflNeo), the floxed allele without Neo (EogtF), and the deleted allele (Eogt−). Eogt exons (closed boxes); Neo and diphtheria toxin (DT3) genes served as positive and negative selection markers, respectively (open boxes); loxP sites (gray triangles); FRT sites (open triangles); KpnI (K), HindIII (H), and SalI (S) restriction sites. Homologous recombination between the WT allele and the targeting vector generated the EogtflNeo allele. The EogtF and Eogt− alleles were obtained by Flp-mediated and Ayu1-Cre-mediated recombination, respectively. Red lines indicate positions of probes used for Southern blotting. Positions of primers used for genotyping are indicated by closed triangles. (B) HindIII- or KpnI/SalI-digested genomic DNA isolated from WT (+/+) or heterozygous floxed neo (+/flNeo) ES cells was analyzed by Southern blotting using the short arm or long arm probe, respectively. In addition to signal from the WT allele (arrow), the flNeo mouse shows an additional band corresponding to the recombinant allele (arrowhead). (C) Genomic DNA isolated from WT, Eogt+/−, and Eogt−/− mice was subjected to genotyping using 3rdloxFw, 3rdloxRv, and 25307Rv primers. (D) Semi-quantitative RT-PCR analysis of total RNA from WT or Eogt−/− brain ECs using primers targeting exons 9 and 11. Minor transcripts lacking exon 10 were detected in Eogt−/− mice (arrow). Gapdh was amplified as an internal control. (E) Quantification of Eogt transcripts in Eogt−/− and WT mouse. qRT-PCR analysis of brain ECs showed a marked decrease in Eogt transcripts. Data are mean ± S.D. from three independent experiments performed in triplicate. Each experiment analyzed pooled total RNA obtained from 10 mice. (F) Lack of EOGT protein expression in Eogt−/− mouse. Lung lysates prepared from adult WT or Eogt−/− mice were analyzed in parallel with cell lysates from HEK293T cells overexpressing Eogt. Immunoblotting was performed using anti-EOGT or β-tubulin antibodies. (G) Whole-mount in situ hybridization for Eogt in the P5 or P15 retina. Vascular staining of Eogt is evident in Eogt+/− retina. Eogt expression in vascular sprouts is indicated by arrows. (H) P5 control Tek-Cre and Tek-Cre EogtF/F retinas were subjected to in situ hybridization. Counter staining with Dylight 594-conjugated IB4 is shown below.
DOI: http://dx.doi.org/10.7554/eLife.24419.011