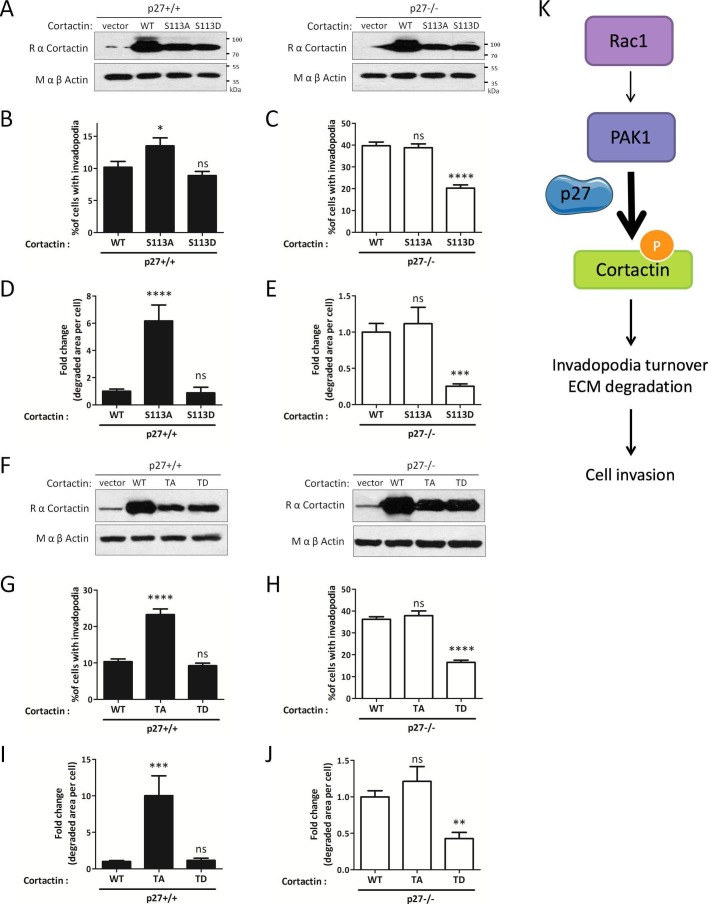
Figure 8. Mimicking phosphorylation of Cortactin restores invadopodia dynamics in p27−/− cells.
(A–E) p27−/− E6 MEFs were infected with empty vector or with vectors encoding wild type Cortactin (WT), S113A-Cortactin (S113A) or S113D-Cortactin (S113D). (A) Cortactin levels after retroviral infection were determined by immunoblot with rabbit anti-Cortactin (H-191) antibodies. β-actin was used as loading control. (B–E) Cells were seeded on gelatin-A488 for 48 hr. After Tks5 staining, cells forming invadopodia (B–C), or the area of degraded gelatin, expressed in fold-change compared to WT Cortactin transfected cells (D–E), were quantified in at least ten fields per condition in each experiment, representing a minimum of 179 cells per genotype. The graphs show the means of at least three independent experiments. (F–J) p27−/− E6 MEFs were infected with empty vector or with vectors encoding WT Cortactin, Cortactin TA (S113A/S150A/S282A) or Cortactin TD (S113D/S150D/S282D). (J) Cortactin levels after retroviral infection were determined as in (A). (G–J) Cells were processed as in (B–E) to quantify cells forming invadopodia (G–H), or the area of degraded gelatin (I–J), with a minimum of 222 cells counted per genotype per experiment. The graphs show the means of 3 independent experiments. (K) Schematic representation of the Rac1/PAK1/phospho-Cortactin pathway involved in invadopodia turnover and matrix degradation and its proposed regulation by p27. ****p<0.0001; ***p<0.001; **p<0.01; *p<0.05.
DOI: http://dx.doi.org/10.7554/eLife.22207.032