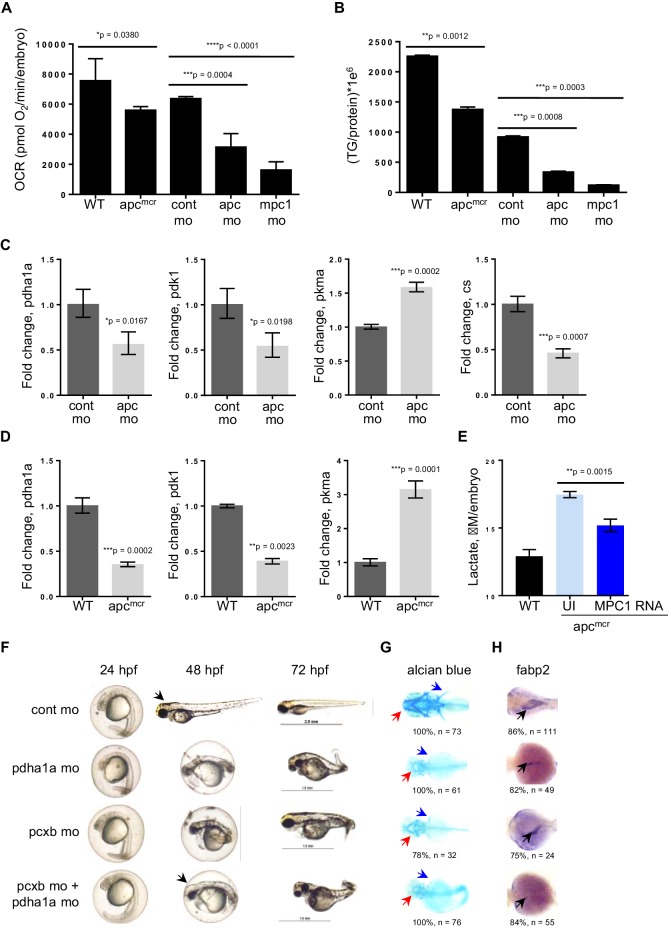
Figure 4. Knock down of mpc1 or apc leads to altered mitochondrial respiration and pyruvate metabolism.
(A) Mitochondrial respiration was evaluated by measuring oxygen consumption rates (OCR) in 72 hpf embryos. (B) Triglyceride (TG) levels were determined in lysates prepared from 72 hpf embryos using a colorimetric assay. (C,D) Quantitative RT-PCR analysis of enzymes involved in pyruvate metabolism in apc mo and (C) apcmcr (D) embryos. pyruvate dehydrogenase alpha 1a (pdha1a); pyruvate dehydrogenase kinase, isozyme 1 (pdk1); pyruvate kinase, muscle, a (pkma); citrate synthase (cs). (E) Lactate levels in apc wild type (WT), un-injected apcmcr (UI) or apcmcr embryos injected with human MPC1 mRNA (MPC1 RNA). For figures A–E, values represent mean ± SD. Graph shown above is representative of at least three independent experiments. Statistical significance was analyzed using unpaired t-test. (F,G,H) Gross phenotype (F), alcian blue staining (G) and in situ hybridization for fabp2 (H) in pdha1a, pcxb, and pcxb + pdha1a mo. pcxb (pyruvate carboxylase b). See also Figure 4—figure supplements 1, 2.
DOI: http://dx.doi.org/10.7554/eLife.22706.010
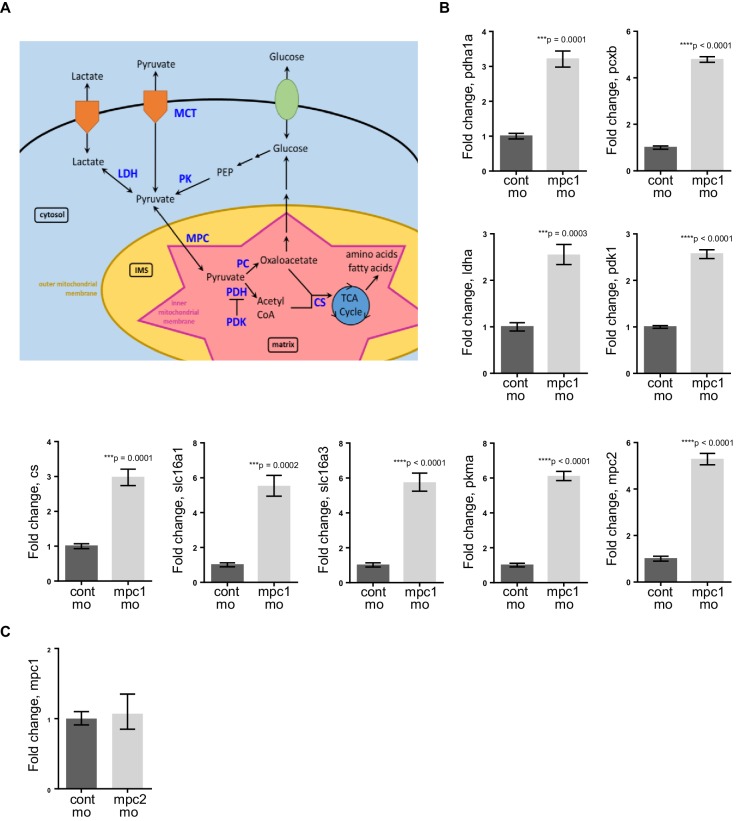
Figure 4—figure supplement 1. Knockdown of mpc1 leads to dysregulated pyruvate metabolism.
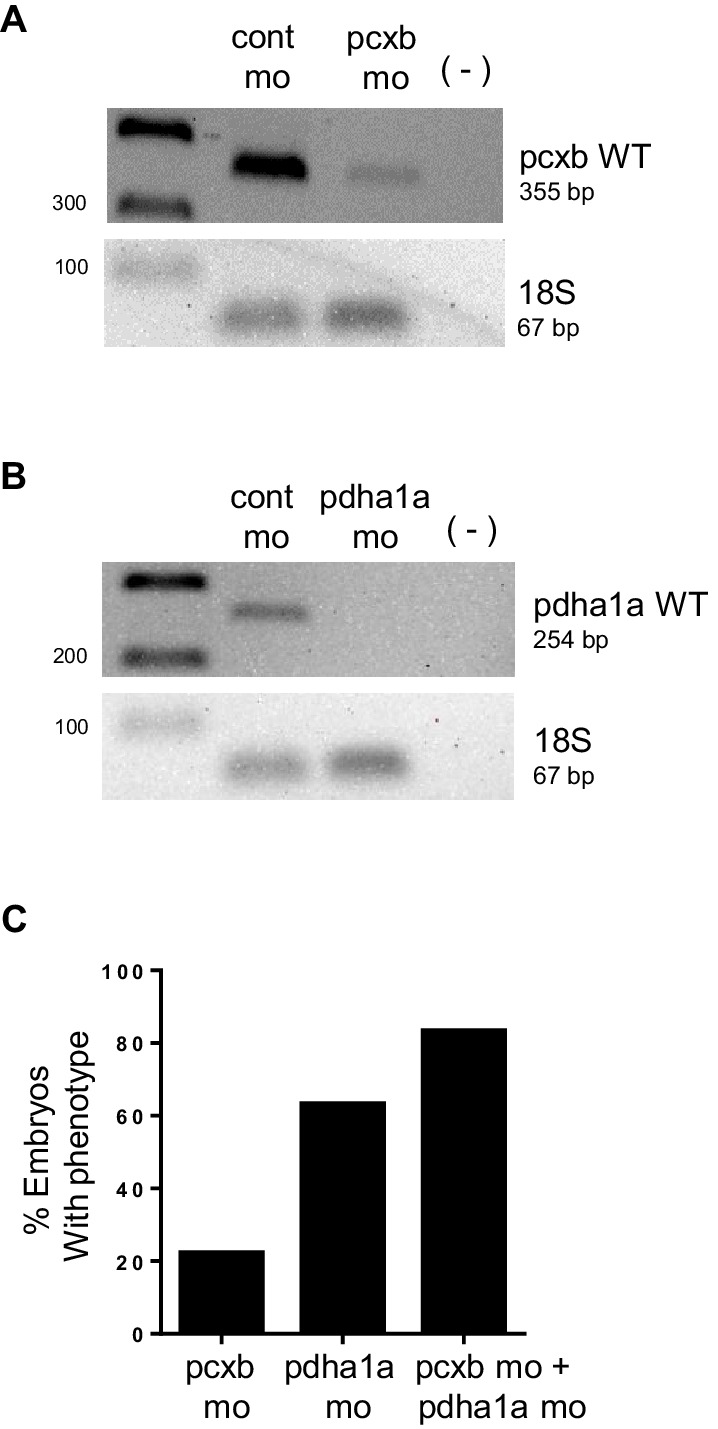
Figure 4—figure supplement 2. PCR analysis confirming knockdown of pdha1a, pcxb.