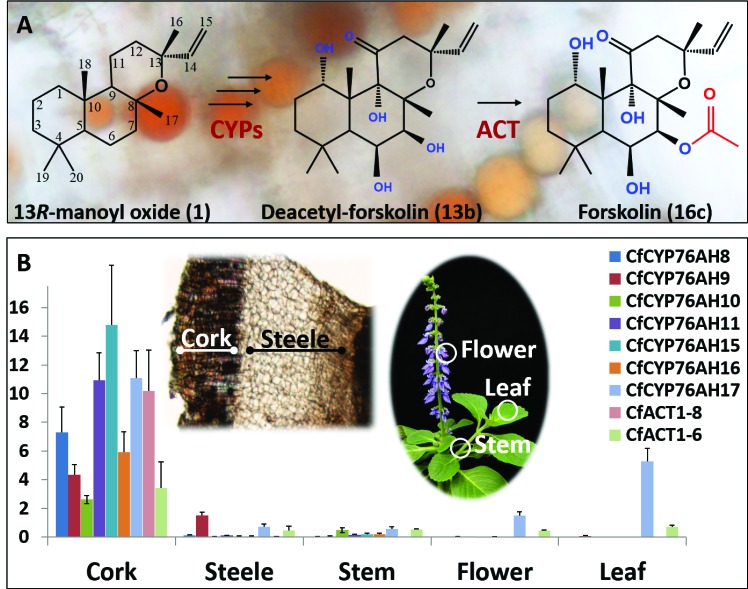
Figure 1. Biosynthesis of forskolin in the root cork cells of C. forskohlii.
(A) Scheme showing the structures of the diterpene precursor 13R-manoyl oxide, deacetylforskolin and forskolin on a background of root cork cells with forskolin containing oil bodies. (B) Transcript profiles of biosynthetic candidate genes in selected tissues of C. forskohlii as shown on the illustrations.
DOI: http://dx.doi.org/10.7554/eLife.23001.003