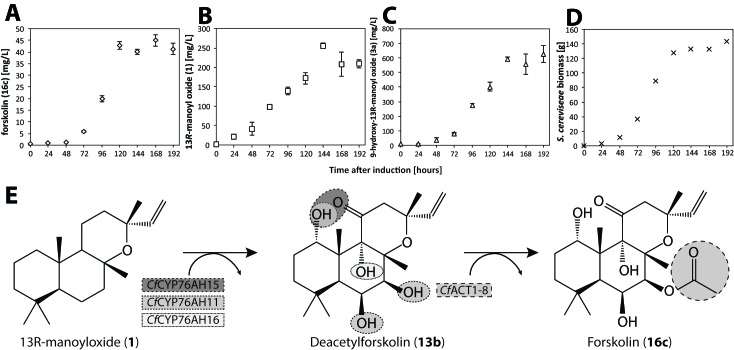
Figure 10. Forskolin production in S. cerevisiae following stable genomic integration of codon-optimized C.forskohlii genes.
(A) Forskolin (16c) accumulation in a fermenter batch using the EVST21543 strain (expressing CfCYP76AH15, CfCYP76HA11, CfCYP76AH16 and CfACT1-8 encoding genes in the EFSC4498 S. cerevisiae strain, optimized for the production of 13R-manoyl oxide [Andersen-Ranberg et al., 2016]). (B) 13R-manoyl oxide (1) accumulation in EVST21543 strain. (C) 9-Hydroxy-13R-manoyl oxide (3a) accumulation in EVST21543 strain. (D) EVST21543 strain biomass monitored during the fermentation process. (E) The biosynthetic pathway used for the production of forskolin in yeast. The fermentation event occurred once, and a triplicate of samples were analysed from each time course.