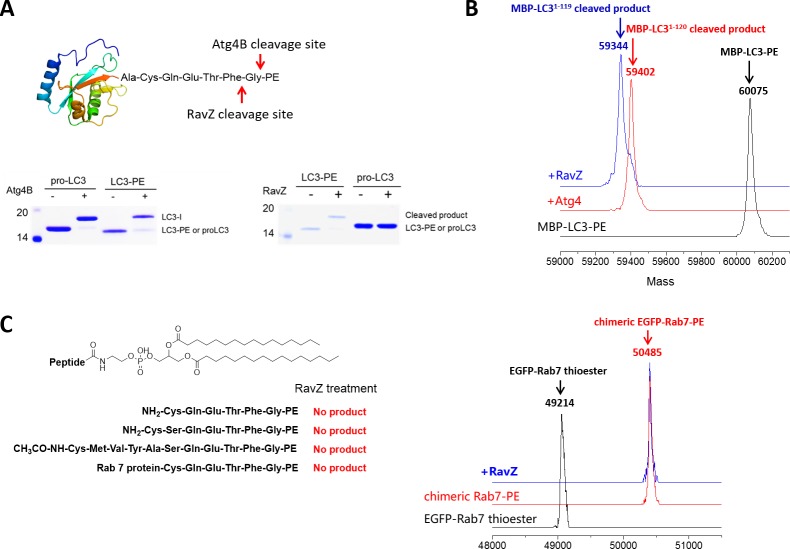
(A) In vitro LC3 cleavage assay by SDS-PAGE. RavZ only cleaves LC3-PE but not pro-LC3, while Atg4B processes both of them. (B) RavZ and Atg4 deconjugate LC3-PE at different sites. ESI-MS spectra of MBP-LC3-PE before and after Atg4B and RavZ treatments were shown. MBP-LC3-PE, Mw calculated 60078, found 60075; MBP-LC31–120, Mw calculated 59391, found 59402; MBP-LC31–119, Mw calculated 59344, found 59344. (C) The C-terminal lipidated peptide of LC3-PE is insufficient for RavZ proteolysis. The C-terminal lipidated peptide of LC3-PE and chimeric PE-modified Rab7 were subjected to RavZ treatment. The lipidated peptides were solved in the HEPES buffer containing detergent (30 mM HEPES 7.4, 50 mM NaCl, 2 mM DTT, 0.1% Triton X-100). RavZ (2 µM) was added to the peptide solution (2 µM) and chimeric PE-modified Rab7 (7 µM). After overnight incubation at 37°C, the reactions were subjected to LC-MS. No cleavage products were observed in these conditions. Right panel shows ESI-MS spectra of EGFP-Rab7 thioester, EGFP-Rab7-PE chimeric protein and RavZ-treated EGFP-Rab7-PE. Chimeric EGFP-Rab7-PE protein, Mw calculated 50500, found 50485.