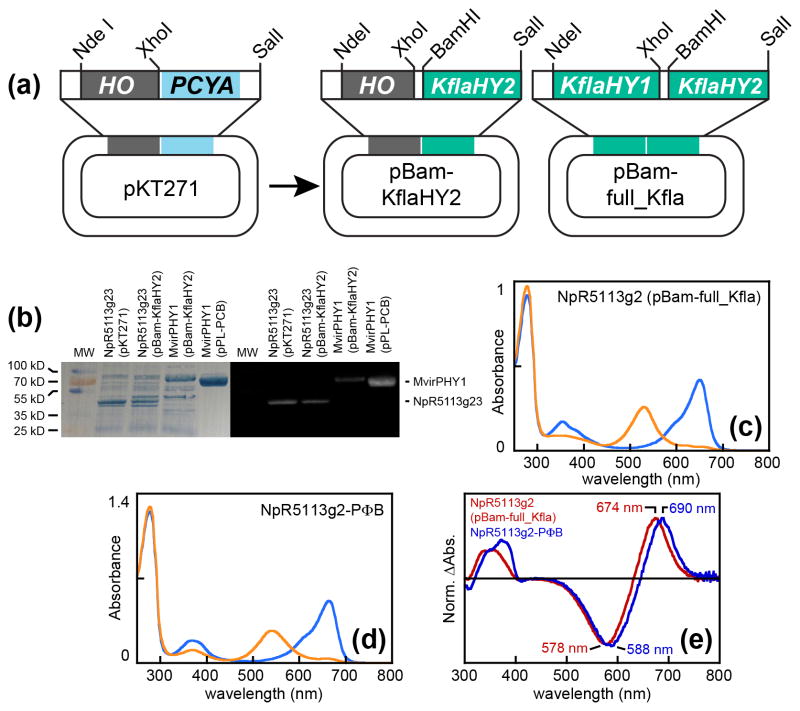
Fig. 4.
Reconstitution of algal bilin biosynthesis in Escherichia coli. (a) Construction of bilin biosynthesis plasmids used in this study is shown. (b) Purified biliprotein photoreceptors are shown after sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (left) and zinc blotting (right) to assess bilin content (Berkelman & Lagarias, 1986). MW, molecular weight standards. (c) Absorption spectra are shown for NpR5113g2 in the 15Z red-absorbing dark state (blue) and 15E green-absorbing photoproduct state (orange) after expression with bilin biosynthesis driven by KflaHY1 and KflaHY2. (d) Absorption spectra are shown for NpR5113g2 in the 15Z red-absorbing dark state (blue) and 15E green-absorbing photoproduct state (orange) after coexpression with biosynthetic machinery for phytochromobilin (PΦB) (Fischer et al., 2005). (e) Normalized photochemical difference spectra are shown for denatured NpR5113g2 incorporating bilin produced using Klebsormidium flaccidum bilin biosynthetic machinery (red trace) or PΦB biosynthetic machinery (dark blue trace), with peak wavelengths indicated.