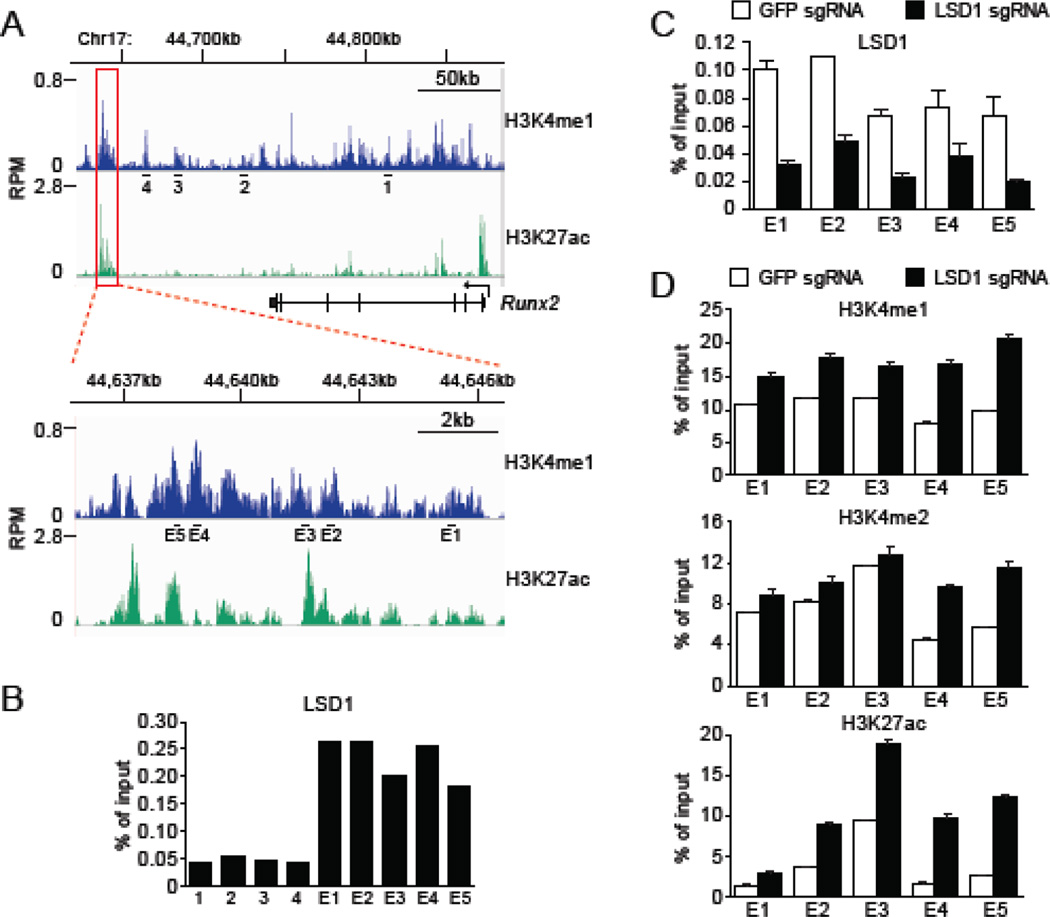
Figure 5.
LSD1 binds to Runx2 downstream enhancer and regulates histone H3 modification. (A) ChIP-seq data for H3K4me1 and H3K27ac at Runx2 locus in C2C12 myoblast. Public data deposited in GEO (GSM1197187 and GSM921130) were analyzed by using SRAtailor software [31]. Candidate enhancer region downstream of the Runx2 gene is indicated by red box and a close up of this region is shown in the lower panel. Black bars and numbers indicate primers used for ChIP-qPCR in B-D. (B, C) ChIP-qPCR analysis with anti-LSD1 antibody at the Runx2 locus. LSD1 enrichment are shown as % signal/input. (B) Cas9-C2C12 were differentiated into myotube in medium containing 2% FBS. (C) LSD1 knockout cells were examined as negative control for ChIP experiment with anti-LSD1 antibody. Error bars indicate SEM from four replicates of qPCR. (D) ChIP-qPCR analysis with indicated antibody at the Runx2 downstream enhancer in LSD1 knockout cells. GFP sgRNA was used as control. Cells were differentiated in medium containing 2% FBS. Results are shown as % signal/input. Error bars indicate SEM from three replicates of ChIP-qPCR.