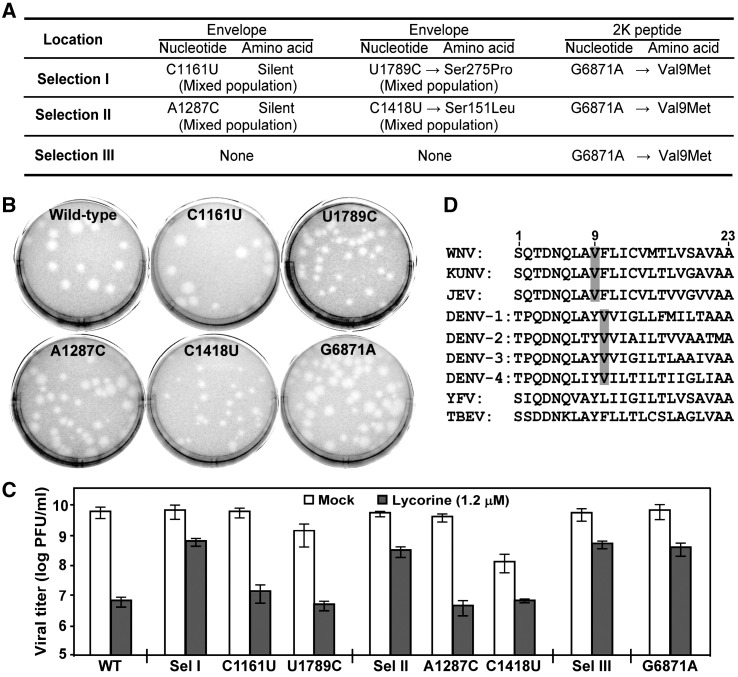
Fig. 6.
Identification of a single-amino acid change in the 2K peptide as a resistance determinant. (A) Summary of mutations identified from the three selections. Locations of the nucleotide and/or amino acid changes are indicated. Mixed populations (containing both the WT-nucleotide and the indicated mutant-nucleotide) were found in the E gene from selections I and II. (B) Plaque morphologies of WT and recombinant C1161U, U1789C, A1287C, C1418U, and G6871A viruses. Plaques were developed in the absence of lycorine. (C) Resistance analyses of WT virus, three independently selected P12 viruses (from Sel, I, II, and III), and recombinant C1161U, U1789C, A1287C, C1418U, and G6871A viruses. The resistance assays were performed as described in the legend to Fig. 5B. (D) Alignment of amino-acid sequences of flavivirus 2K peptide. The 23-amino acid sequences of 2K peptide are aligned for nine flaviviruses. The conserved Val residues in viruses from JEV- and DENV-serocomplexes are shaded in grey. The 2K peptide sequences of WNV, KUNV, JEV, DENV-1, DENV-2, DENV-3, DENV-4, YFV and TBEV are derived from GenBank accession numbers AF404756, D00246, AF315119, U88535, M29095, M93130, AY947539, X03700, and AF069066, respectively.