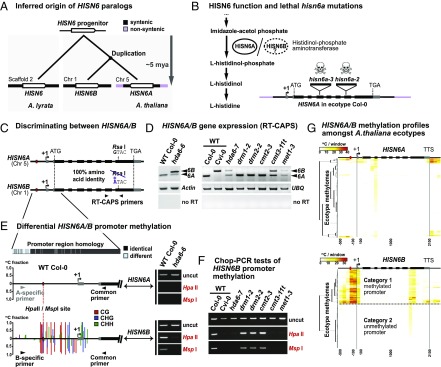
Fig. 1.
HDA6, CMT3, and MET1 silence HISN6B via promoter region DNA methylation. (A) Inferred phylogenetic origin of HISN6 paralogs based on synteny between HISN6B flanking regions on A. thaliana chromosome 1 and sequences including the single copy HISN6 gene of A. lyrata scaffold 2 (Fig. S1 A and B). (B, Left) HISN6A/B protein function in histidine biosynthesis. Steps upstream of Imidazole-acetol phosphate are omitted. (B, Right) Gene structure of HISN6A in ecotype Col-0: UTRs, exons, and T-DNA insertion positions in mutant alleles are indicated by gray boxes, black boxes, and inverted triangles, respectively. (C) RT-CAPS assay for discrimination of HISN6A and HISN6B mRNAs using primers flanking a polymorphic RsaI site present only in HISN6A. (D) HISN6A/B expression analysis via RT-CAPS in hda6-6, hda6-7, drm1-2, drm2-2, cmt2-3, cmt3-11t, and met1-3 mutants compared with WT Col-0 or Cvi-0. Actin and ubiquitin (UBQ) reactions served as loading controls. Reactions omitting reverse transcriptase (no RT) are controls for genomic DNA contamination. (E) Analysis of DNA methylation in HISN6A and HISN6B promoter regions. Bar plots show WT Col-0 methylation profiles, color-coded by sequence context (CG, CHG, and CHH), tabulated as fractional cytosine methylation (y-axis), based on methylome data of Stroud et al. (40). No methylation was detected in the HISN6A promoter. Gel images show Chop-PCR assays of cytosine methylation status at a HISN6A/B promoter HpaII/MspI site (red dotted line) in WT Col-0 or the hda6-6 mutant. Reactions omitting restriction enzymes (uncut) demonstrate equivalent DNA input. HISN6A/B primer specificity was verified by sequencing PCR products. (F) Analysis of HISN6B promoter methylation in the mutant series of D, using Chop-PCR. (G) Hierarchical clustering of cytosine methylation profiles for HISN6A (Top) or HISN6B (Bottom) genes in 892 ecotype methylomes of A. thaliana. Methylated cytosines, based on data of Kawakatsu et al. (41), were tallied within 100-bp nonoverlapping windows starting 500 bp upstream of the transcription start site (+1) and stopping 300 bp downstream of the TTS.