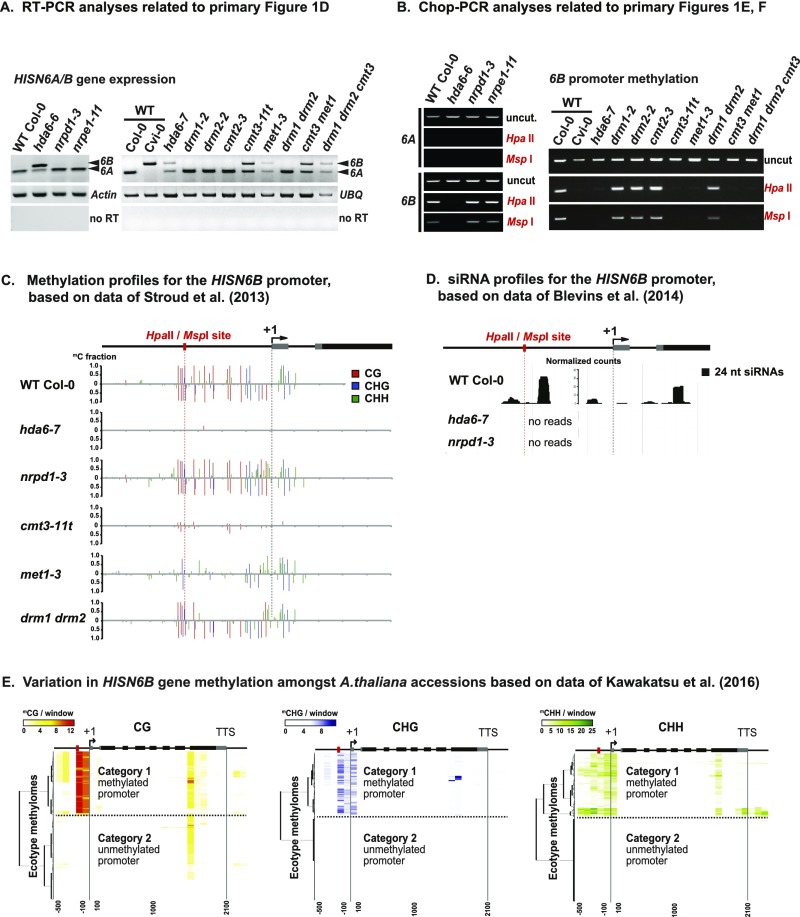
Fig. S2.
HISN6A and HISN6B expression and methylation in WT Col-0 and in mutants affecting chromatin modification and gene silencing. (A) RT-PCR products were digested with RsaI to discriminate HISN6A/B expression in WT Col-0 or Cvi-0 and the indicated Col-0 mutants (complete panel corresponding to Fig. 1D). (B) HISN6A and HISN6B promoter methylation in WT and various Col-0 mutants was analyzed by Chop-PCR at the promoter region HpaII/MspI restriction site (complete panel corresponding to Fig. 1 E and F). Reactions omitting restriction enzymes (uncut) control for equivalent DNA input in all reactions. (C) Cytosine methylation profiles are shown for a 600-bp region encompassing the HISN6B promoter and transcription start site (+1). Methylomes of 3-wk-old leaves isolated from WT and mutant lines of the Col-0 ecotype were analyzed [data from Stroud et al. (40)]. Bars represent fractional methylation per cytosine position, color-coded by sequence context (CG, CHG, and CHH). The HpaII/MspI restriction enzyme site used for Chop PCR assays is highlighted by a red dotted line. (D) A genome browser shot showing small RNA profiles of 2-wk-old leaves of WT, hda6-7, or nrpd1-3 mutants of the Col-0 ecotype. Read counts corresponding to 24-nt siRNAs are plotted in black on the y-axis, normalized by total mapped reads [data from Blevins et al. (15)]. (E) Hierarchical clustering analysis of HISN6A/B methylation, as in Fig. 1G, but divided by CG, CHG, or CHH sequence context [data from Kawakatsu et al. (41)].