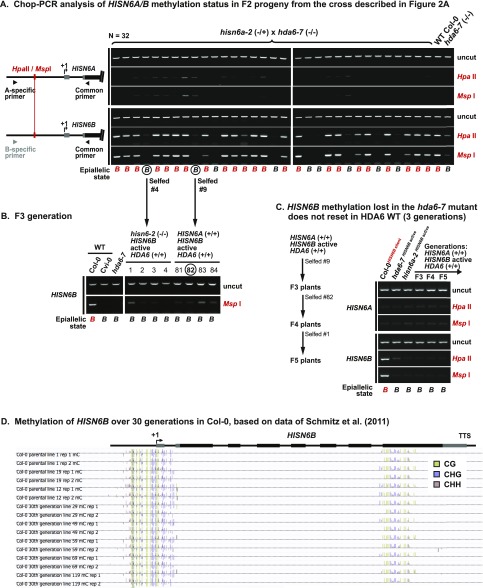
Fig. S3.
Stable HISN6B methylation is lost in hda6 mutants and not regained upon restoration of WT HDA6 genes. (A) Chop-PCR DNA methylation analysis of F2 progeny from the cross in Fig. 2A testing HISN6A/B methylation at the promoter HpaII/MspI site. Reactions omitting restriction enzymes serve to control for equivalent DNA input to all reactions (uncut). PCR specificity was verified by sequencing uncut PCR products. (B) Chop-PCR analysis of HISN6B promoter methylation in F3 progeny from the cross in Fig. 2A. (C) Chop-PCR analysis of HISN6A/B promoter methylation in a HISN6B active lineage derived from an hda6 mutant background, in three self-fertilized generations after restoration of HDA6 function (F3, F4, and F5 generation from the cross in Fig. 2A). WT Col-0 (HISN6B silent) and hda6-7 and hisn6a-2 mutants (both HISN6B active) served as controls. (D) Browser shot of HISN6B methylation profiles in WT Col-0 lines propagated by single-seed descent for 30 generations. Profiles of independent parental lines and 30th generation lines are shown, with the methylcytosine sequence context (CG, CHG, or CHH) indicated using different colors. Data are from Schmitz et al. (44), displayed using the genome browser tool described in that paper (neomorph.salk.edu/30_generations/browser.html).