Significance
By culturing a human colorectal cancer (CRC) cell line (HCA-7) in 3D, we have generated two cell lines (CC and SC) with distinct morphological, genetic, biochemical, and functional properties. Using this 3D system, we have discovered that increased tyrosine phosphorylation of MET and RON results in cetuximab resistance in the SC cell line that can be overcome by addition of the dual MET/RON tyrosine kinase inhibitor, crizotinib. We have also identified that increased epithelial, but not stromal, versican staining correlates with reduced survival in a clinically annotated CRC tissue microarray.
Keywords: colorectal cancer, versican, HPGD, 3D culture, cetuximab resistance
Abstract
We previously reported that single cells from a human colorectal cancer (CRC) cell line (HCA-7) formed either hollow single-layered polarized cysts or solid spiky masses when plated in 3D in type-I collagen. To begin in-depth analyses into whether clonal cysts and spiky masses possessed divergent properties, individual colonies of each morphology were isolated and expanded. The lines thus derived faithfully retained their parental cystic and spiky morphologies and were termed CC (cystic) and SC (spiky), respectively. Although both CC and SC expressed EGF receptor (EGFR), the EGFR-neutralizing monoclonal antibody, cetuximab, strongly inhibited growth of CC, whereas SC was resistant to growth inhibition, and this was coupled to increased tyrosine phosphorylation of MET and RON. Addition of the dual MET/RON tyrosine kinase inhibitor, crizotinib, restored cetuximab sensitivity in SC. To further characterize these two lines, we performed comprehensive genomic and transcriptomic analysis of CC and SC in 3D. One of the most up-regulated genes in CC was the tumor suppressor 15-PGDH/HPGD, and the most up-regulated gene in SC was versican (VCAN) in 3D and xenografts. Analysis of a CRC tissue microarray showed that epithelial, but not stromal, VCAN staining strongly correlated with reduced survival, and combined epithelial VCAN and absent HPGD staining portended a poorer prognosis. Thus, with this 3D system, we have identified a mode of cetuximab resistance and a potential prognostic marker in CRC. As such, this represents a potentially powerful system to identify additional therapeutic strategies and disease-relevant genes in CRC and possibly other solid tumors.
Traditionally, epithelial cells have been cultured on plastic as a flat monolayer, precluding formation of their characteristic apico-basolateral structural organization. Pioneering work by Mina Bissell and Joan Brugge has shown that select breast epithelial cell lines can be grown in 3D in Matrigel as polarizing cysts with intact apico-basolateral polarity (1–3). These 3D cultures have been used to study oncogene-induced transformation and are shown to better predict in vivo behavior than 2D cultures (4, 5). Similar work in colonic epithelial cells has lagged behind; a notable exception is the work of Alan Hall and colleagues with Caco-2 cells that form uniform polarizing cysts in 3D Matrigel (6).
We sought to identify human colorectal cancer (CRC) lines that exhibit apico-basolateral polarity in a better-defined 3D environment than Matrigel, which is a complex, incompletely defined extracellular matrix secreted by Engelbreth–Holm–Swarm mouse sarcoma cells (7). We previously observed that a human CRC line, HCA-7, cultured in type-1 collagen, gave rise to colonies consisting of unilamellar cysts with intact apico-basolateral polarity or less frequent colonies composed of irregular solid masses of cells (8). We derived CC and SC lines from cystic and spiky colonies, respectively. When injected subcutaneously into athymic nude mice, CC formed well-differentiated, encapsulated tumors, whereas SC formed poorly differentiated, locally invasive tumors (8).
Here, we provide an in-depth morphological, genetic, biochemical, and functional characterization of CC and SC. Despite appearing virtually indistinguishable on plastic and on Transwell filters, CC and SC exhibit dramatic morphological and functional differences in 3D.
Results
Isolation and Propagation of CC and SC from Parental HCA-7.
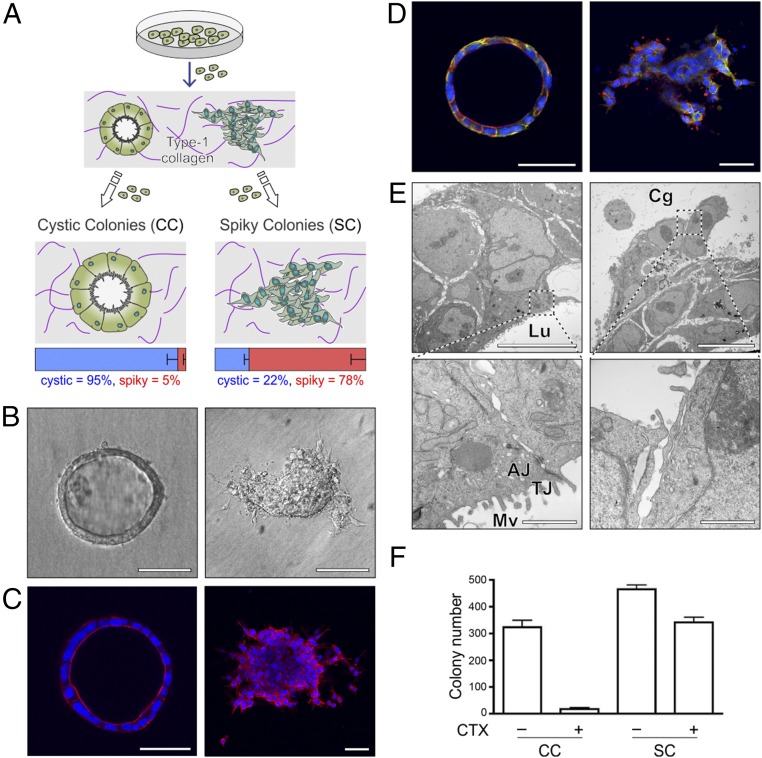
In an effort to identify human CRC lines that polarized in a well-defined extracellular matrix, we cultured a number of human CRC cell lines (HCA-7, Caco-2, SK-CO15, HCT-8, LS174T, SW480, HCT-116, and RKO) in type-1 collagen. After 15 d of culture in type-I collagen, only HCA-7 formed colonies with smooth-edged, single-layered cysts with hollow cavities (Fig. 1 A and B, Left). Upon closer inspection, less frequent colonies were also observed that appeared to be solid masses with protrusions (Fig. 1 A and B, Right). We manually isolated individual cystic and spiky colonies and propagated them on plastic. CC and SC lines thus derived maintained their distinctive cystic and spiky morphologies, respectively, upon repeated passage in 3D. CC and SC were scored for their morphological appearance after 15 d in 3D type-1 collagen; 95% of CC appeared cystic and 78% of SC appeared spiky (Fig. 1A).
Fig. 1.
Generation and characterization of HCA-7–derived CC and SC. (A) A human CRC cell line, HCA-7, exhibited two distinct morphologies when grown in 3D in type-1 collagen: hollow unilamellar cystic colonies (CC) and spiky colonies (SC) that formed solid masses with protrusions into the collagen matrix. Individual CC and SC colonies were manually isolated and expanded on plastic. CC and SC maintained their characteristic appearance upon repeated passage in 3D. Individual colonies from CC and SC 3D cultures were scored for cystic or spiky morphology. Results are displayed as the mean fraction of total colonies (%) ± SEM. (B) DIC images of representative CC (Left) and SC (Right) colonies. (Scale bars: 50 µm.) (C) Confocal images through the equatorial plane of representative colonies from CC and SC clones stained for DAPI (blue) and phalloidin (red). (Scale bars: 50 µm.) (D) Confocal images through the equatorial plane of representative CC and SC colonies stained for EGFR (blue) and ezrin (red). (Scale bars: 50 µm.) (E) Representative CC and SC colonies were fixed and processed for transmission electron microscopy as described in Materials and Methods. (Top, Left and Right) A section of colonies with CC- and SC-like morphology, respectively. Lu, the luminal (apical) side; Cg, the collagen (basal) side of cysts. Both colonies are displayed in identical orientation. (Scale bars: 2 µm.) (Lower panels) Higher magnifications from each morphology. AJ, adherens junction; Mv, microvilli; tight junction, TJ. Note the lack of TJ in the magnified region corresponding to the protrusions in SC colonies. (Scale bars: 500 nm.) (F) Two thousand cells were cultured in type-1 collagen for 17 d. Fresh medium was added with or without cetuximab (CTX, 3 μg/mL) every 2–3 d. Colony count was determined using a GelCount plate reader. Results are plotted as mean counts ± SEM. P values from a two-tailed unpaired t test are 0.0003 for CC and 0.0049 for SC.
We next performed a more detailed morphological characterization of CC and SC. Representative differential interference contrast (DIC) images in Fig. 1A show that CC colonies form hollow unilamellar cysts and that SC colonies form solid masses with protrusions (Fig. 1B). These findings are further confirmed by confocal images through the equatorial planes of CC and SC colonies stained for nuclei and F-actin (Fig. 1C). In addition, apico-basolateral polarity was maintained in CC colonies as determined by subapical ezrin immunoreactivity and EGFR decorating the lateral membranes (Fig. 1D). Transmission electron microscopy of CC colonies showed that apico-basolateral architecture was maintained with rudimentary microvilli detected at the apical surface with tight junctions (TJ) and adherens junctions (AJ) at nearby cell–cell junctions (Fig. 1E, Left panels). Within the protruding regions of SC colonies, TJ and AJ were not observed (Fig. 1E, Right panels). In contrast to their marked morphological differences in 3D, both CC and SC exhibited a similar epithelial cobble-stone appearance in 2D (SI Appendix, Fig. S1A), although SC grew faster than CC (SI Appendix, Fig. S1C). Similarly, both CC and SC formed uniform polarized monolayers on Transwell filters with a similar basolateral localization of CDH1 and ITGB1 (SI Appendix, Fig. S1B).
Identification of a Mode of Cetuximab Resistance.
We previously reported that CC form well-differentiated tumors in nude mice, whereas SC form poorly differentiated, locally invasive tumors (8). We next sought to determine if there were additional functional differences between CC and SC. EGFR-neutralizing monoclonal antibodies (cetuximab and paintumumab) are approved by the US Food and Drug Administration for the treatment of advanced wild-type KRAS CRCs (9). Because HCA-7 cell lines are wild type for KRAS, we tested the efficacy of cetuximab against CC and SC in 3D. Fig. 1F shows that cetuximab has a greater inhibitory effect on the growth of CC compared with SC, whereas it has only a modest effect on the growth of either CC or SC in 2D (SI Appendix, Fig. S1D).
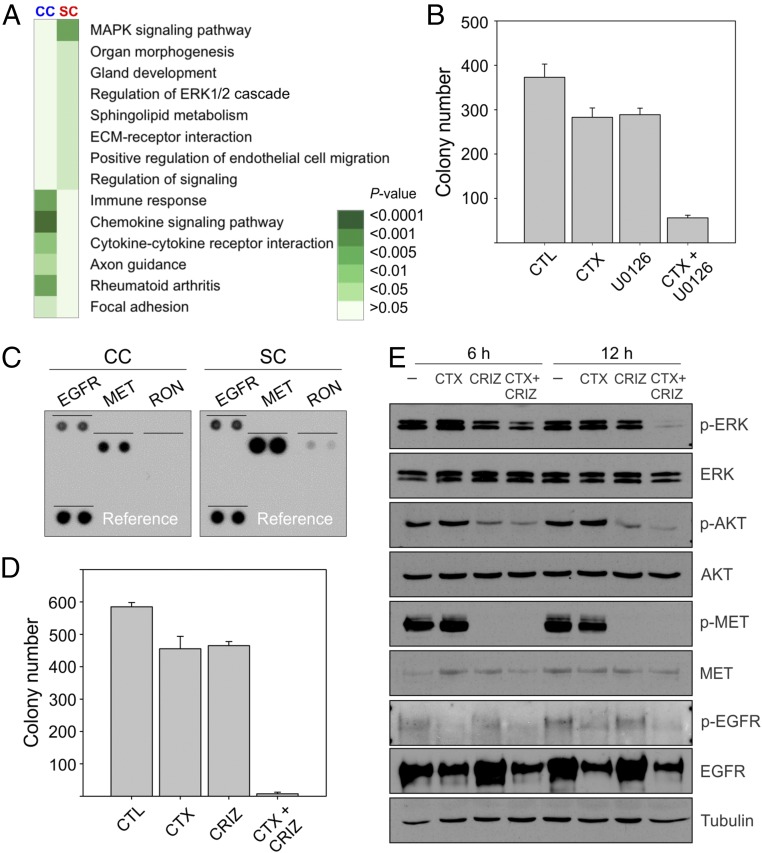
We set out to determine the mechanism(s) underlying cetuximab resistance in SC. There were no known genetic events that could explain the resistance to cetuximab in SC; KRAS, BRAF, NRAS, PIK3CA, and EGFR were wild type, and MET was not amplified (SI Appendix, Table S1). Nor did we detect any differences between CC and SC at the level of EGFR mRNA expression, total and cell-surface EGFR protein, and EGFR activation following EGF stimulation (SI Appendix, Table S2 and Fig. S2 A and B). Based on Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of gene expression, we found that MAPK signaling was the most up-regulated signaling pathway in SC compared with CC (Fig. 2A and Fig. 3 B and C). Bardelli and coworkers recently showed that addition of a MEK inhibitor (pimasertib) to cetuximab could prevent cetuximab resistance (10). Consistent with that result, addition of another MAPK inhibitor (U0126) to cetuximab in SC results in cooperative growth inhibition (Fig. 2B). To determine upstream signaling pathways that might be responsible for increased MAPK signaling in SC, we performed a receptor tyrosine kinase (RTK) array on CC and SC and found that there was a selective increase in phosphorylated MET and RON in SC compared with CC (Fig. 2C). This led us to examine the effect of crizotinib, a dual MET/RON tyrosine kinase inhibitor. Whereas neither cetuximab nor crizotinib alone significantly inhibited the growth of SC, the combination markedly inhibited growth (Fig. 2D). As expected, crizotinib decreased MET tyrosine phosphorylation and cetuximab decreased total and tyrosine-phosphorylated EGFR (Fig. 2E). The combination of cetuximab and crizotinib led to a marked reduction in ERK1/2 and AKT phosphorylation (Fig. 2E).
Fig. 2.
Mechanism of cetuximab resistance in SC. (A) KEGG analysis of gene expression shows that MAPK signaling is the most up-regulated pathway in SC versus CC. (B) Two thousand SC cells were cultured in type-1 collagen for 2 wk. Fresh medium was added with cetuximab (CTX, 3 μg/mL) and/or the MEK inhibitor U0126 (1 µM) every 2–3 d as indicated. Colony count are plotted as mean ± SEM P < 0.05 for the combined treatment compared with control and individual treatments (two-tailed unpaired t test). (C) A portion of the RTK array showing relative levels of phosphorylation of EGFR, MET, and RON between CC and SC. Each signal is spotted in duplicate; reference is spotted on the lower left. (D) Two thousand SC cells were cultured in type-1 collagen for 2 wk. Fresh medium was added with cetuximab (CTX, 3 μg/mL) and/or the MET/RON inhibitor crizotinib (CRIZ, 0.05 µM) every 2–3 d as indicated. Colony counts are plotted as mean ± SEM P < 0.05 for the combined treatment compared with control and individual treatments (two-tailed unpaired t test). (E) One hundred thousand SC cells were cultured in type-1 collagen for 5 d and incubated with cetuximab (CTX, 3 μg/mL) and/or the MET/RON inhibitor crizotinib (CRIZ, 0.05 µM) for 6 or 24 h as indicated. Middle-collagen-layer–containing cells were lysed, resolved on SDS/PAGE, and immunoblotted for the indicated proteins and their phosphorylation state.
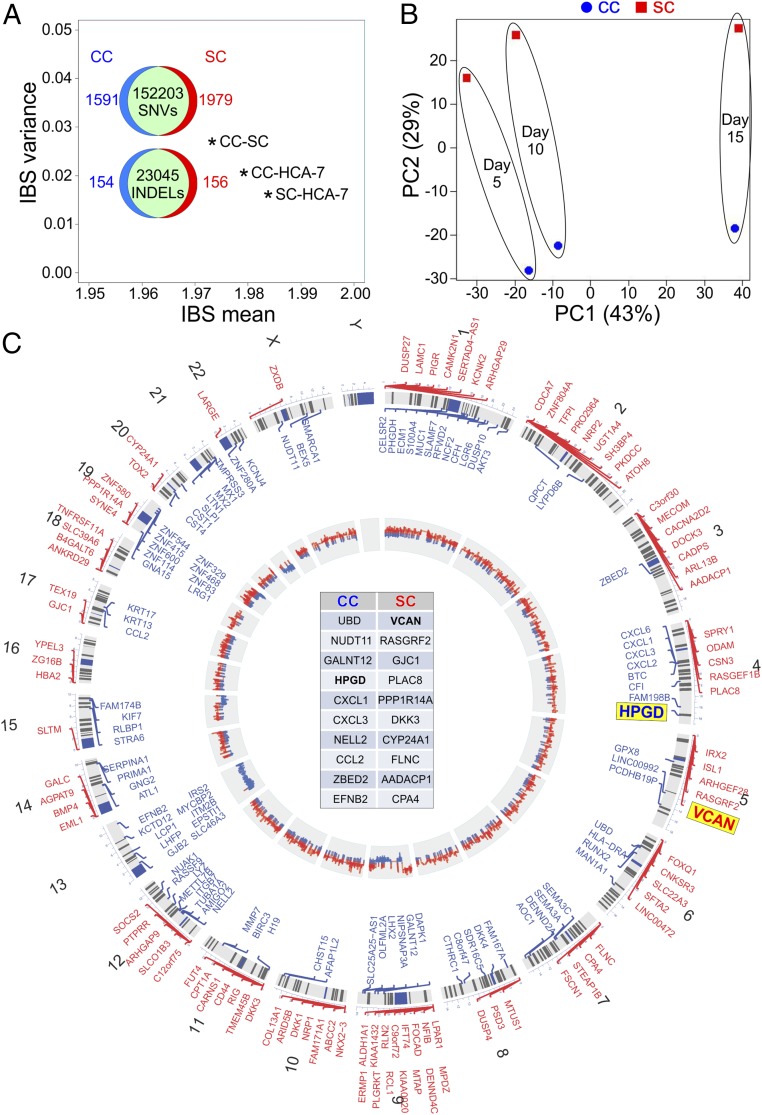
Fig. 3.
Genetic and transcriptional characterization of CC and SC. (A) Genetic relatedness among CC, SC, and parental HCA-7 by pairwise IBS distance plot. A comparison of SNVs and INDELs between CC and SC is shown within the IBS plot. (B) Principal component analysis of transcriptional profiles for CC (blue) and SC (red) cultured in 3D for 5, 10, and 15 d, represented by small, medium, and large spheres, respectively. (C) Circos plot of copy number variation (CNV) and differential expression between CC and SC. Chromosomal location is marked on the outer circle. The chromosomal location of the top 100 up-regulated genes in SC (red) and CC (blue) is depicted. The top 10 up-regulated genes in CC and SC are displayed in the center table in decreasing order of expression. The innermost circle shows relative CNV between CC and SC with gains in CC and SC depicted as blue and red, respectively.
Genetic Characterization of CC and SC.
Based on the morphological and functional differences observed between CC and SC, we decided to perform a more detailed characterization of these lines. CC and SC are derived from microsatellite unstable HCA-7 (11). Microsatellite unstable tumors are unable to repair base-pair mismatches and have hundreds to thousands of mutations. We performed whole-exome sequencing of parental HCA-7 and their CC and SC derivatives. We identified single nucleotide variants (SNVs) and insertions and deletions (INDELs) for each cell line and calculated pairwise identity-by-state (IBS), in which two individuals are observed to have zero, one, or two alleles in common at a given locus (12). The IBS of all three pairs (CC-SC, CC-HCA-7, and SC-HCA-7) exhibited a mean close to 2 and minor variance, confirming the genetic relatedness of these cell lines (Fig. 3A). Among the three pairs, CC-SC had the lowest IBS mean and largest variance, indicating that CC and SC were more different from each other than parental HCA-7. Nevertheless, there were 152,203 common SNVs and 23,045 INDELs shared between CC and SC, consistent with their common parental origin (Fig. 3A). All three lines shared a homozygous frameshift mutation in type 2 TGF-β receptor (TGFBR2), resulting in receptor truncation, a homozygous mutation in APC resulting in a premature stop at codon 1554 and a heterozygous R150W mutation in TP53. There were no nonsynonymous mutations in mismatch repair genes (PMS2, MLH1, MSH2, and MSH6). However, HCA-7, CC, and SC all exhibited hypermethylation of MLH1, and MLH1 protein was not detected by immunohistochemistry (IHC) (SI Appendix, Fig. S3). All three lines were wild type for KRAS, BRAF, PIK3CA, and EGFR.
The genetic differences between CC and SC accounted for less than 2% of total SNVs and less than 1% of total INDELs (Venn diagrams in Fig. 3A). There were heterozygous missense mutations in 156 genes in CC and 172 genes in SC (SI Appendix, Fig. S4A). Of note, there were 14 genes with homozygous mutations unique to SC (SI Appendix, Table S1 and Fig. S4A). In CC, six genes had unique homozygous missense mutations (SI Appendix, Table S1 and Fig. S4A). In addition, CC and SC had five and four unique insertions or deletions, respectively (SI Appendix, Table S1).
We next examined gene expression in CC and SC in 3D culture. RNA was isolated from CC and SC cultured in 3D for 5, 10, and 15 d for microarray analysis. Principal component analysis showed that both cell type and time contributed to the transcriptional landscape (Fig. 3B). To focus on the contribution of cell type rather than time in culture, we focused on genes with consistent differential expression between CC and SC at all three time points. The top 100 genes (blue for CC, red for SC) from that list are aligned along their respective chromosomes in the Circos plot in Fig. 3C (complete list in SI Appendix, Table S2). The top 10 genes for each line are listed in the center of the Circos plot in decreasing order of fold-change.
The inner circle of the Circos plot depicts copy-number gains and losses as determined by exome sequencing. Based on CC and SC genetic comparison, we found copy-number gain in chromosome 9p24.3–9p21.3 in SC that may underlie the consistent overexpression of a cohort of genes in this region. Similarly, there were gene clusters showing copy number gains and losses in the inner circle with corresponding changes in gene expression in the outer circle. The most striking examples were chromosomes 13 and 21 where nearly all of the chromosomes showed high-copy-number variation (CNV) and expression in CC. Chromosome and digital karyotyping established that most of the CNV and overexpression clusters could be explained by gains or losses of whole chromosomes or parts of chromosomes, for example, loss of one chromosome 13 in SC and gain of two additional chromosome 21’s in CC (SI Appendix, Fig. S5 and Table S3). Beyond that, we found little evidence suggesting a contribution of copy-number changes to the expression differences between CC and SC. Although there were no global differences in DNA methylation between CC and SC (SI Appendix, Fig. S6), there were differences in methylation of specific histone H3 residues visible by immunostaining in both 3D cultures and tumors in nude mice (SI Appendix, Fig. S7).
In three recent CRC gene expression studies (CCS, CRCA, and CCMS), each study identified a subtype associated with a low degree of differentiation, epithelial-to-mesenchymal transition, and poor prognosis; these subtypes were 3 (48 genes), 5 (185 genes), and 4 (288 genes), respectively (13–15). We found that these gene signatures are selectively up-regulated in SC, consistent with their less differentiated and more invasive features compared with CC (SI Appendix, Fig. S4B).
In Vivo and in Vitro Validation of HPGD and VCAN Expression in CC and SC Cells.
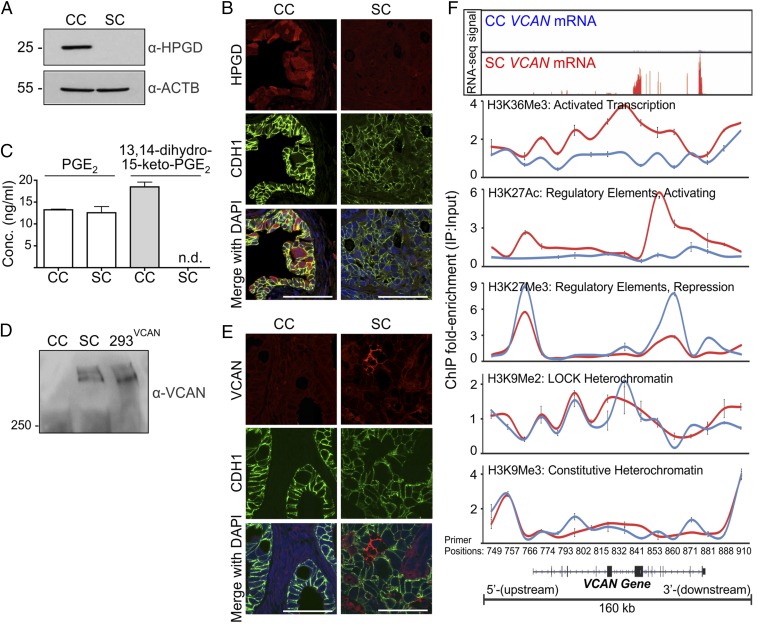
We decided to focus on selected genes up-regulated in CC and SC. The top four up-regulated genes in CC are UBD, GALNT12, NUDT11, and HPGD. Based on our previous work in parental HCA-7 cells linking EGFR and eicosanoid metabolism, we elected to study HPGD, a key enzyme in prostaglandin metabolism (16). HPGD metabolizes PGE2 to 13,14-dihydro-15-keto-PGE2, thereby acting as a tumor suppressor (17, 18). We detected HPGD protein in cell lysates from CC, but not SC (Fig. 4A). In addition, the HPGD metabolite 13,14-dihydro-15-keto-PGE2 was measured in conditioned medium from CC, but not SC (Fig. 4C). Moreover, HPGD immunofluorescence was observed in the epithelial component of CC xenografts in athymic nude mice, but not in their SC counterparts (Fig. 4B). Thus, CC overexpress the tumor suppressor HPGD, which is consistent with their more differentiated and less invasive features. However, incubation with the HPGD inhibitor, SW033291, did not result in increased growth or spiky morphologic conversion of CC colonies. Instead, at higher concentrations (100 µM) CC colonies showed reduced size, indicating arrest of colony growth (SI Appendix, Fig. S8 A and B).
Fig. 4.
In vitro and in vivo validation of HPGD and VCAN expression in CC and SC. (A) CC and SC grown in 3D in type-1 collagen for 15 d were lysed and resolved on 10% SDS/PAGE and immunoblotted for HPGD and β-actin. (B) Formalin-fixed, paraffin-embedded (FFPE) sections from CC and SC tumors established as subcutaneous xenografts in athymic nude mice were immunostained for HPGD (red), CDH1 (green), and DAPI (blue). (Scale bars: 50 µm.) (C) LC/MS/MS analysis of PGE2 and the HPGD metabolite (13,14-dihydro-15-keto-PGE2) from the conditioned medium of CC and SC cultured in 3D for 14 d. Results are plotted as mean concentration (ng/mL) ± SEM. n.d., not detected. (D) Conditioned media from CC and SC grown in 3D and VCAN-overexpressing HEK293Ts cultured on plastic were treated with chondroitinase, resolved on 7% SDS/PAGE, and immunoblotted for VCAN. (E) FFPE sections from CC and SC xenografts were immunostained for VCAN (red), CDH1 (green), and DAPI (blue). (Scale bars: 50 µm.) (F) ChIP with antibodies against the indicated histone modifications was performed on chromatin isolated from CC (blue) and SC (red) colonies grown in 3D in type-1 collagen for 15 d (n = 6). The immunoprecipitated DNA was amplified with 16 real-time PCR primers spaced across the VCAN locus as indicated at the bottom of the panels. Results represent ChIP fold-enrichment over input as mean ± SD. Top panel represents the matched RNA-Seq signal for VCAN gene expression in CC and SC.
VCAN was the most up-regulated gene in SC compared with CC when placed in a 3D type-1 collagen environment (Fig. 3C). VCAN is a large chondroitin sulfate proteoglycan that is produced by both cancer cells and tumor stroma. It is part of a stromal gene signature that recently has been linked to poor clinical outcome in individuals with CRC (19, 20). In addition to its overexpression in SC, VCAN protein was detected in conditioned medium from SC, but not CC (Fig. 4D). Likewise, VCAN immunofluorescence was observed in the epithelial component of SC xenografts in athymic nude mice, but not in CC xenografts (Fig. 4E).
Transcriptional regulation of VCAN has been previously described (21, 22). Because the VCAN locus was not amplified or deleted in either CC or SC, we considered whether the chromatin state across the VCAN locus might contribute to the regulation of VCAN. H3K9Me2 and H3K27Me3 are dynamic modifications that are broadly enriched within large (50 kb–10 Mb) facultative heterochromatin domains that have been termed large organized chromatin lysine(K)-modified heterochromatin domains (LOCKs) (23–25), nuclear lamin-associated domains (26), and partially (DNA) methylated domains (25). These domains are known to acquire DNA hypomethylation early in CRC progression compared with normal tissue, and it is thought that loss of DNA methylation in these regions might confer regulatory plasticity to these domains through dynamic regulation of chromatin modifications (27, 28). Indeed, we noted that the VCAN locus was present within a large heterochromatin domain that is DNA hypomethylated in CRC (29).
We next performed ChIP assays of SC and CC cultured in 3D to test whether reprogrammed chromatin modifications were specifically targeted to VCAN chromatin. We examined multiple chromatin modifications using a set of 16 pairs of quantitative PCR primers spaced at ∼10-kb intervals across the 160-kb VCAN locus, including regions upstream and downstream from the genic portion. As expected, the interior of this region was depleted for H3K9Me3 and enriched for H3K9Me2 in both CC and SC, consistent with a CRC facultative heterochromatin domain that is common to both (Fig. 4F, CC in blue, SC in red). We further observed enrichment of H3K27Me3 at specific regions within this domain in CC (Fig. 4F). This modification is strongly linked to transcriptional repression and gene silencing, consistent with low VCAN expression in CC. In contrast, these same regions were depleted of H3K27Me3 and highly enriched with H2K27Ac in SC. H3K27Ac is typically enriched at gene regulatory elements, including enhancers, to encode a chromatin state permissive for transcriptional activation. Indeed, we observed strong enrichment of the transcription-coupled modification H3K36Me3 over the VCAN gene in SC, but not in CC, consistent with transcriptional up-regulation (30). Thus, the VCAN locus resided within a large chromatin domain that acquired reciprocal activating and repressive chromatin modifications in SC versus CC, suggesting an epigenetic basis for overexpression of VCAN in SC.
Analysis of VCAN and HPGD Immunoreactivity in a CRC Tissue Microarray.
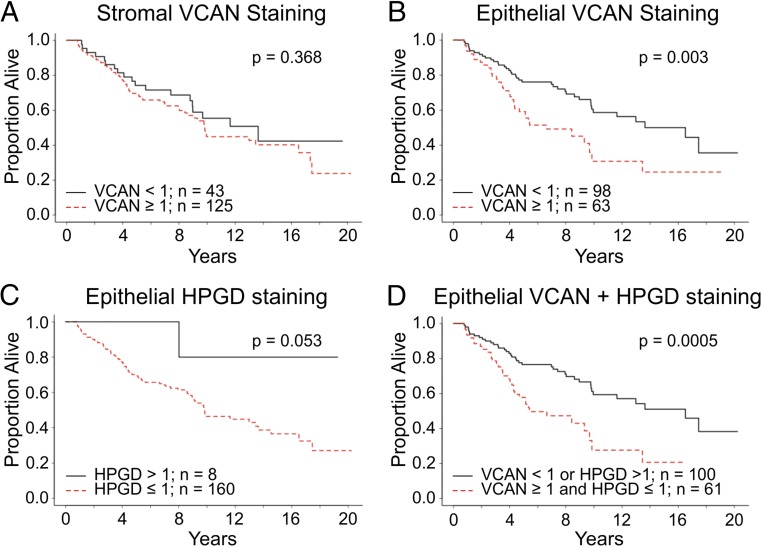
We next examined VCAN and HPGD immunoreactivity in a clinically annotated tissue microarray consisting of 174 CRCs from 174 individuals (8). There was no difference in overall survival for individuals with high and low staining for VCAN in the stroma (Fig. 5A). However, individuals with high staining for VCAN in the epithelium had significantly worse overall survival than those with low epithelial VCAN staining (Fig. 5B and SI Appendix, Fig. S9A). HPGD immunoreactivity was detected only in the epithelium (SI Appendix, Fig. S9B). Loss of HPGD staining was frequently observed in CRCs, with only eight tumors displaying appreciable staining (Fig. 5C). There was a trend for individuals with HPGD-positive tumors to have a better outcome. Individuals with tumors lacking HPGD and high VCAN staining had a worse survival than those with only high epithelial VCAN immunoreactivity (Fig. 5D). Thus, this 3D system has allowed us to identify that epithelial, and not stromal, VCAN staining portends a poor prognosis in CRC. In separate analyses we found that increased HPGD mRNA expression correlated with better survival and that increased VCAN expression was detected in the poor-prognosis, subtype C CRC samples (SI Appendix, Fig. S10 A and B) (31–33).
Fig. 5.
Kaplan–Meier plots of overall survival by biomarker classification based on a tissue microarray of 174 CRCs from 174 individuals. (A) Overall survival comparison by IHC staining of stromal VCAN (log rank P = 0.368). (B) Overall survival comparison by IHC staining of epithelial VCAN (log rank P = 0.003). (C) Overall survival comparison by IHC staining of epithelial HPGD (log rank P = 0.053). (D) Overall survival comparison between two groups separated by IHC staining of epithelial VCAN and HPGD. Black line indicates cumulative survival of individuals with high-VCAN and low-HPGD tumors; red line indicates individuals with low-VCAN and high-HPGD tumors (log rank P = 0.0005).
Discussion
Here, we provide a detailed genomic and transcriptomic characterization of a 3D model system to study CRC. By placing single HCA-7 cells into a well-defined extracellular matrix (type-1 collagen), we have generated two cell lines (CC and SC) with contrasting morphologic and functional properties. Of note, CC and SC appear indistinguishable when cultured on plastic and on Transwell filters. A number of other CRC lines also were cultured in type-1 collagen, but only HCA-7 exhibited a uniform cystic morphology.
A striking functional difference between CC and SC was the fact that CC were markedly growth-inhibited by cetuximab in 3D, whereas SC were refractory to its growth inhibitory effects. Neither line responded to cetuximab in 2D. We excluded all known genetic events that could explain the resistance of SC to cetuximab. Both CC and SC cells were wild type for KRAS, BRAF, PIK3CA, EGFR, and MET. Moreover, cetuximab resistance in SC could not be explained by differential EGFR cell-surface availability; EGF led to equivalent EGFR tyrosine phosphorylation, and the extracellular domain antibody, C225, showed equivalent surface staining in CC and SC (SI Appendix, Fig. S2 A and B). When comparing the gene expression profile of CC and SC cells by KEGG analysis, we observed increased MAPK signaling in SC cells (Fig. 2A). Moreover, a small-molecule MAPK inhibitor, U0126, in combination with cetuximab, led to cooperative growth inhibition of SC in 3D (Fig. 2B).
To find upstream kinases mediating MAPK activation, we performed an RTK array and found a selective increase in MET and RON tyrosine phosphorylation (Fig. 2C). Although MET amplification has been reported as a mechanism of acquired resistance to EGFR inhibition (34, 35), increased tyrosine phosphorylation of MET (independent of its amplification) has not. Crizotinib is a broadly acting, small-molecule tyrosine kinase inhibitor that is approved for clinical use for individuals with non–small-cell lung carcinoma (36). Although crizotinib is particularly effective in tumors with ALK and ROS1 mutations, it has the highest affinity for MET with RON being an additional target (37–39). Crizotinib, in combination with cetuximab, led to marked growth inhibition of SC cells in 3D (Fig. 2D) Consistent with the reduction in colony number, the cetuximab/crizotinib combination also worked cooperatively to reduce downstream signaling as monitored by levels of ERK1/2 and AKT phosphorylation (Fig. 2E). Finally, as to the likely source of increased MET/RON tyrosine phosphorylation, among other possibilities, up-regulation of MET/RON ligands (HGF/HGFL) and/or inactivation of MET/RON tyrosine phosphatases may play a regulatory role (40, 41).
We performed whole-exome sequencing of HCA-7, CC, and SC. Pairwise analysis among the three revealed that the CC-SC pair was the most divergent. Nevertheless, there was more than 98% similarity between CC and SC as scored by SNVs and INDELs (Fig. 3A). The SNVs and INDELs unique to CC and SC are listed in SI Appendix, Table S1. Although the majority of clustered changes in gene expression could be explained by losses or gains in chromosomal regions as determined by karyotyping, the overexpression of VCAN and HPGD was not due to copy-number gains or losses or chromosomal rearrangements (SI Appendix, Fig. S5 and Table S3). Future studies will be needed to determine if any of these genetic events in SC contribute to cetuximab resistance.
We compared CC and SC gene expression over time in 3D by microarray. Two of the top four genes overexpressed in CC are bona fide tumor suppressor genes—GALNT12 and HPGD—whereas the role of the other two—UBD and NUDT11—in CRC is uncertain (42–44). Because we previously showed EGFR-induced COX-2 expression and basolateral release of one of the COX-2 products, PGE2 in HCA-7 cells, we decided to focus on HPGD because HPGD metabolizes PGE2 to 13,14-dihydro-15-keto-PGE2 (16, 45, 46). HPGD thus reduces the levels of PGE2, which is thought to be the major tumor-promoting eicosanoid (47). There was a marked reduction in intestinal tumors when ApcMin mice were crossed to Cox-2 null mice and a marked increase in tumor burden when ApcMin mice were crossed to Hpgd null mice, supporting the notion that tumor suppression in intestinal neoplasia is achieved by inhibiting PGE2 production (via Cox-2 loss) or assuring its degradation (via Hpgd) (48). We now report that the less aggressive HCA-7–derived CC show up-regulation of HPGD compared with the more aggressive SC and also report selective up-regulation of the HPGD metabolite, 13,14-dihydro-15-keto-PGE2, in CC. Among the EGFR ligands, AREG potently induces COX-2, so it will be of interest to examine the effect of the EGFR-signaling axis on HPGD expression (49). However, when directly tested, HPGD inhibition did not confer a transformed phenotype to CC colonies as assessed by colony growth and morphologic conversion, indicating that HPGD may not be functionally linked to the benign CC phenotype (SI Appendix, Fig. S8 A and B). On the other hand, HPGD expression remained strongly associated with better survival in CRC (Fig. 5 C and D).
VCAN was the most up-regulated gene in SC. Of interest, VCAN has also been shown to interact with and influence EGFR signaling (50, 51). VCAN is a major component of the extracellular matrix, being produced by both tumor cells and the surrounding stroma (52). Recent work has identified VCAN as part of a poor prognosis stromal gene signature (19, 20). However, Michael Karin and coworkers demonstrated that VCAN was produced by epithelial-derived Lewis lung cancer cells and that it was a potent activator of macrophages, resulting in proinflammatory tumor progression (53). We show that VCAN is selectively overexpressed in the less differentiated, more invasive SC compared with their CC counterparts. In a clinically well-annotated CRC tissue microarray, we show that epithelial, but not stromal, VCAN immunoreactivity is linked to poor prognosis in CRC. Another group, using a different antibody to VCAN, reached a different conclusion as to the prognostic significance of epithelial versus stromal VCAN staining (54). Future studies are needed to determine whether VCAN produced by epithelial or stromal cells differs in form or function. Our current studies do not address the biological function of VCAN. It is a large protein with a number of splice and proteolytic isoforms that bind to cell-surface and extracellular proteins (55, 56). Our work identifies epigenetic regulation of VCAN expression (21, 22).
The present studies do not provide an explanation for the persistent morphological and functional differences between CC and SC in 3D. When we isolate individual spiky colonies from SC type-1 collagen cultures and embed them as single cells in type-1 collagen, we consistently observe a predominant spiky morphology in the resulting colonies. A similar experiment with the infrequent CC-derived spiky colonies results in colonies with predominantly cystic morphology. This Luria–Delbrück fluctuation-like analysis is insufficient to elucidate the genetic and/or epigenetic versus environmental basis of the phenotypic differences between CC and SC (57). However, because both CC and SC form predominantly cystic colonies in Matrigel, we favor the notion that exposure to specific environments, in this case type-1 collagen, provides cues that magnify the underlying genetic and epigenetic differences between the two lines. Whatever the ultimate mechanism(s) underlying differences between CC and SC in 3D, this system provides a useful tool to identify epithelial-derived, disease-relevant genes, as well as additional therapeutic strategies in CRC.
Materials and Methods
Reagents.
PureCol bovine type-1 collagen was purchased from Advanced Biomatrix. All cell culture components were purchased from HyClone Laboratories. Protein G agarose and rhodamine–phalloidin were purchased from Life Technologies. Anti-ezrin antibody was purchased from Cell Signaling (#3145).
Cell Culture.
All cell lines were maintained in DMEM containing 10% (vol/vol) bovine growth serum, nonessential amino acids, l-glutamine, and penicillin–streptomycin. For 3D cultures, type-1 collagen was diluted at 2 mg/mL in DMEM containing 10% (vol/vol) FBS. Briefly, assays were set up using three collagen layers, 400 µL each, in 12-well culture dishes, with the middle layer containing the single-cell suspension at 5,000 cells/mL. Medium (400 µL) with or without reagents was added on top and changed every 2–3 d. Colonies were observed and counted after 14–17 d.
Isolation of CC and SC.
Individual cystic or spiky colonies from 20-d-old HCA-7 collagen cultures were picked and trypsinized for 15–20 min at room temperature (RT) and transferred to individual wells of a six-well dish containing complete medium and expanded. Four cystic clones (CC1-4) and two spiky clones (SC1-2) were isolated. CC3 and SC1 clones, used throughout this study and designated CC and SC, respectively, have maintained their distinct morphologies in 3D culture for over 20 passages.
Microarray Analysis.
RNA from the middle layer of CC and SC collagen cultures was extracted using TRIzol Reagent (Life Technologies). The RNeasy Mini Kit (Qiagen) was used to clean up 50–100 μg of each total RNA sample. Six samples were analyzed on Affymatrix 133 Plus 2.0 array: CC and SC (5, 10, and 15 d). All samples were 3× diluted (1.1 µL H2O, 2.2 µL sample, 3.3 µL 20 mM Tris). Quality controls were performed on a Nanodrop 2000 and Bioanalyzer 2100. After successful hybridization and scanning, Robust Multichip Average (RMA) was performed to normalize the six samples. Consistent differences between SC and CC were computed by combining time points from the same cell type together.
Immunoblotting.
The middle collagen layer was removed and placed into a 100 µL RIPA buffer for 30 min at 4 °C. Lysates were precleared by centrifugation at 14,000 × g for 10 min. Supernatants were diluted 1:1 with 2× Laemmli buffer [5% (vol/vol) β-mercaptoethanol], boiled for 5 min, and resolved on 8% SDS/PAGE.
VCAN immunoblotting.
HEK293T cells expressing recombinant versican V1 (plasmid gift from Dieter Zimmermann, Department of Pathology, University of Zurich, Zurich) served as a positive control. Conditioned medium from CC, SC, and positive controls was treated with 0.5 U/mL ABC chondroitinase (Sigma) in 100 mM sodium acetate, 50 mM Tris⋅HCl, pH 8, buffer for 1 h at 37 °C. Digestion was terminated by addition of nonreducing Laemmli sample buffer, and samples were run on a 4–20% gradient gel.
Histone immunoblotting.
Three-dimensional cultures were collagenase-treated (1% collagenase in complete medium) at 37 °C for 1 h. Cells were collected by centrifugation, washed twice with PBS, and lysed in Nonidet P-40 buffer containing protease inhibitors (complete Protease Inhibitor Mixture Tablets from Roche) and phosphatase inhibitors (PhosSTOP Phosphatase Inhibitor Mixture Tablets from Roche). Histones were extracted with H2SO4 and resolved on SDS/PAGE (58). All gels were subsequently processed as described previously (59).
Immunofluorescence.
3D IF.
Collagen sandwich was fixed in 4% (wt/vol) paraformaldehyde for 30 min at RT. Middle layer was removed and placed into IF buffer (1% BSA, 1% Triton X-100 in PBS) overnight. Before wash 568-phalloidin and DAPI were added for 4 h at 4 °C.
IF.
Primary antibodies were added at 1:500 overnight in IF buffer. Samples were then washed, and secondary antibodies were added at 1:1,000 for 4 h at 4 °C. Samples were washed and whole-mounted beneath a #1.5 glass coverslip supported with spacers.
Tissue section.
Tumor xenografts were fixed in neutralized formalin and embedded in paraffin. Slices were deparaffinized with serial histoclear and ethanol. Antigen retrieval was performed in citrate buffer (pH 6) with high pressure for 30 min. Primary antibodies used were the following: anti-VCAN (DSHB, 12C5; 1:100); anti-HPGD (LS-BIO, 2C10; 1:100); anti–E-cadherin (Abcam, EP700Y; 1:500), and DAPI. Secondary antibodies from Life Technologies (AlexaFluor-linked) were used. Slides were washed and mounted in Prolong (Life Technologies). Confocal microscopy was performed using a Nikon A1R.
Immunohistochemistry.
Tumor xenografts were fixed in neutralized formalin and embedded in paraffin. Slices were deparaffinized with serial histoclear and ethanol. Antigen retrieval was performed in citrate buffer (pH 6) with high pressure for 20 min. Primary antibodies used were the following: anti-H3K9Me2 (1:3,000, mouse antibody); anti-H3K9Me3 (1:1,000, rabbit antibody); and anti-H3K27Me3 (1:200, mouse antibody). Secondary antibodies from Dako were used.
Colony Counting.
Colonies were counted using GelCount (Oxford Optronix) with identical acquisition and analysis settings and represented as mean from triplicates ± SEM. For cystic and spiky morphology, counts were performed manually from three individual wells and represented as mean ± SEM.
Transmission Electron Microscopy.
Collagen cultures were rinsed with 0.1 M sodium cacodylate buffer, fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4 at RT for 1 h and stored at 4 °C overnight. Samples were washed in cacodylate buffer and incubated with 1% osmium tetroxide for 1 h at RT, followed by additional washing with 0.1 M cacodylate buffer. Subsequently, the samples were dehydrated through a graded series of ethanol washes, followed by two exchanges of pure propylene oxide (PO). Samples were infiltrated with Epon 812 resin and PO in a 1:3 ratio for 30 min at RT, followed by a 1:1 ratio for 1 h at RT, and stored overnight at RT. The samples were subsequently infiltrated with resin for 48 h and allowed to polymerize at 60 °C for 48 h. Thick sections (0.5–1 µm) were collected using a Leica Ultracut microtome, contrast-stained with 1% toluidine blue, and imaged with a Nikon AZ100 microscope. Ultra-thin sections (70–80 nm) were cut, collected on 300-mesh copper grids, and poststained with 2% uranyl acetate and then with Reynold’s lead citrate. Samples were subsequently imaged on the Philips/FEI Tecnai T12 electron microscope at various magnifications.
Human RTK Array.
Cell lysates from 3D culture were collected and protein concentration in each sample was measured by the BCA assay (Thermo Scientific). Three hundred milligrams of protein were analyzed using the Human Phospho-Receptor Tyrosine Kinase Array Kit (R&D Systems) according to the manufacturer’s protocol. After applying chemiluminescence detection solution, membranes were exposed to imaging film and developed using a Kodak X-Omat processor (Kodak).
Statistical Analyses.
Two-tailed, two-sample t tests were used to determine statistical significance. P values of less than 0.05 were considered significant. Calculations were performed using GraphPad and R-2.15. DNA methylation data were analyzed with Lumi package for R on the original intensity data (.idat files) (60). Microarray data were normalized with RMA, followed by analyses in R and Excel. Functional enrichment analysis on the up-regulated and down-regulated genes was implemented separately in a Gene Ontology biological process as well as in KEGG pathways by WebGestalt (61, 62). Enrichment P values were generated by a hypergeometric test and adjusted by Benjamini and Hochberg’s multiple test (63). Gene set enrichment analysis was performed using software provided by software.broadinstitute.org/gsea/index.jsp.
Supplementary Material
Acknowledgments
We thank Hengtao Liu and Jinyang Zhang for assistance with karyotyping; Janice A. Williams for help with the transmission electron microscopy; Emily J. Poulin for critical review of the manuscript; and Emily and Mac Brown for their generous support. We also acknowledge the support of Vanderbilt University’s Cell Imaging, Translational Pathology, and Flow Cytometry Shared Resources. This work was supported by National Cancer Institute (NCI) Grant RO1 CA 46413 (to R.J.C.) and NCI Grant P50 95103 from Gastrointestinal Special Programs of Research Excellence (to M.K.W. and R.J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618297114/-/DCSupplemental.
References
- 1.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4(4):359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 3.Hughes CS, Postovit LM, Lajoie GA. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10(9):1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 4.Martin KJ, Patrick DR, Bissell MJ, Fournier MV. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS One. 2008;3(8):e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simian M, Bissell MJ. Organoids: A historical perspective of thinking in three dimensions. J Cell Biol. 2017;216(1):31–40. doi: 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaffe AB, Kaji N, Durgan J, Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J Cell Biol. 2008;183(4):625–633. doi: 10.1083/jcb.200807121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleinman HK, Martin GR. Matrigel: Basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15(5):378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Li C, et al. Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer. J Clin Invest. 2014;124(5):2172–2187. doi: 10.1172/JCI71103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothenberg ML, et al. Randomized phase II trial of the clinical and biological effects of two dose levels of gefitinib in patients with recurrent colorectal adenocarcinoma. J Clin Oncol. 2005;23(36):9265–9274. doi: 10.1200/JCO.2005.03.0536. [DOI] [PubMed] [Google Scholar]
- 10.Misale S, et al. Vertical suppression of the EGFR pathway prevents onset of resistance in colorectal cancers. Nat Commun. 2015;6:8305. doi: 10.1038/ncomms9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkland SC. Dome formation by a human colonic adenocarcinoma cell line (HCA-7) Cancer Res. 1985;45(8):3790–3795. [PubMed] [Google Scholar]
- 12.Goh L, et al. Assessing matched normal and tumor pairs in next-generation sequencing studies. PLoS One. 2011;6(3):e17810. doi: 10.1371/journal.pone.0017810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Sousa E Melo F, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 14.Sadanandam A, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marisa L, et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coffey RJ, et al. Epidermal growth factor receptor activation induces nuclear targeting of cyclooxygenase-2, basolateral release of prostaglandins, and mitogenesis in polarizing colon cancer cells. Proc Natl Acad Sci USA. 1997;94(2):657–662. doi: 10.1073/pnas.94.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Backlund MG, et al. 15-Hydroxyprostaglandin dehydrogenase is down-regulated in colorectal cancer. J Biol Chem. 2005;280(5):3217–3223. doi: 10.1074/jbc.M411221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Y, Tong M, Liu S, Moscow JA, Tai HH. NAD+-linked 15-hydroxyprostaglandin dehydrogenase (15-PGDH) behaves as a tumor suppressor in lung cancer. Carcinogenesis. 2005;26(1):65–72. doi: 10.1093/carcin/bgh277. [DOI] [PubMed] [Google Scholar]
- 19.Calon A, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47(4):320–329. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 20.Isella C, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47(4):312–319. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 21.Rahmani M, et al. Versican: Signaling to transcriptional control pathways. Can J Physiol Pharmacol. 2006;84(1):77–92. doi: 10.1139/y05-154. [DOI] [PubMed] [Google Scholar]
- 22.Ricciardelli C, Sakko AJ, Ween MP, Russell DL, Horsfall DJ. The biological role and regulation of versican levels in cancer. Cancer Metastasis Rev. 2009;28(1-2):233–245. doi: 10.1007/s10555-009-9182-y. [DOI] [PubMed] [Google Scholar]
- 23.Hawkins RD, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6(5):479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41(2):246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hon GC, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22(2):246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453(7197):948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 27.Pujadas E, Feinberg AP. Regulated noise in the epigenetic landscape of development and disease. Cell. 2012;148(6):1123–1131. doi: 10.1016/j.cell.2012.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Schell MJ, et al. A composite gene expression signature optimizes prediction of colorectal cancer metastasis and outcome. Clin Cancer Res. 2016;22(3):734–745. doi: 10.1158/1078-0432.CCR-15-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schell MJ, et al. A multi-gene mutation classification of 468 colorectal cancers reveals a prognostic role for APC. Nature Comm. 2016;7:11743. doi: 10.1038/ncomms11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B, et al. NCI CPTAC Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513(7518):382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 35.Bean J, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104(52):20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw AT, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368(25):2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 38.Shaw AT, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371(21):1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui JJ, et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK) J Med Chem. 2011;54(18):6342–6363. doi: 10.1021/jm2007613. [DOI] [PubMed] [Google Scholar]
- 40.Sangwan V, et al. Regulation of the Met receptor-tyrosine kinase by the protein-tyrosine phosphatase 1B and T-cell phosphatase. J Biol Chem. 2008;283(49):34374–34383. doi: 10.1074/jbc.M805916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trusolino L, Bertotti A, Comoglio PM. MET signalling: Principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11(12):834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 42.Guda K, et al. Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc Natl Acad Sci USA. 2009;106(31):12921–12925. doi: 10.1073/pnas.0901454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan DW, et al. Ubiquitin D is correlated with colon cancer progression and predicts recurrence for stage II-III disease after curative surgery. Br J Cancer. 2010;103(7):961–969. doi: 10.1038/sj.bjc.6605870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grisanzio C, et al. Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proc Natl Acad Sci USA. 2012;109(28):11252–11257. doi: 10.1073/pnas.1200853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, et al. TISSUE REGENERATION. Inhibition of the prostaglandin-degrading enzyme 15-PGDH potentiates tissue regeneration. Science. 2015;348(6240):aaa2340. doi: 10.1126/science.aaa2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan M, et al. 15-Hydroxyprostaglandin dehydrogenase inactivation as a mechanism of resistance to celecoxib chemoprevention of colon tumors. Proc Natl Acad Sci USA. 2009;106(23):9409–9413. doi: 10.1073/pnas.0902367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10(3):181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oshima M, et al. Suppression of intestinal polyposis in Apc delta716 knockout mice by inhibition of cyclooxygenase 2 (COX-2) Cell. 1996;87(5):803–809. doi: 10.1016/s0092-8674(00)81988-1. [DOI] [PubMed] [Google Scholar]
- 49.Cheng JC, Fang L, Chang HM, Sun YP, Leung PC. hCG-induced Sprouty2 mediates amphiregulin-stimulated COX-2/PGE2 up-regulation in human granulosa cells: A potential mechanism for the OHSS. Sci Rep. 2016;6:31675. doi: 10.1038/srep31675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Damasceno KA, et al. HER-2 and EGFR mRNA expression and its relationship with versican in malignant matrix-producing tumors of the canine mammary gland. PLoS One. 2016;11(8):e0160419. doi: 10.1371/journal.pone.0160419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee HC, et al. Cancer metastasis and EGFR signaling is suppressed by amiodarone-induced versican V2. Oncotarget. 2015;6(40):42976–42987. doi: 10.18632/oncotarget.5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agrawal D, et al. Osteopontin identified as lead marker of colon cancer progression, using pooled sample expression profiling. J Natl Cancer Inst. 2002;94(7):513–521. doi: 10.1093/jnci/94.7.513. [DOI] [PubMed] [Google Scholar]
- 53.Kim S, et al. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature. 2009;457(7225):102–106. doi: 10.1038/nature07623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Wit M, et al. Lumican and versican are associated with good outcome in stage II and III colon cancer. Ann Surg Oncol. 2013;20(Suppl 3):S348–S359. doi: 10.1245/s10434-012-2441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du WW, Yang W, Yee AJ. Roles of versican in cancer biology: Tumorigenesis, progression and metastasis. Histol Histopathol. 2013;28(6):701–713. doi: 10.14670/HH-28.701. [DOI] [PubMed] [Google Scholar]
- 56.Wu YJ, La Pierre DP, Wu J, Yee AJ, Yang BB. The interaction of versican with its binding partners. Cell Res. 2005;15(7):483–494. doi: 10.1038/sj.cr.7290318. [DOI] [PubMed] [Google Scholar]
- 57.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2(6):1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 59.Singh B, Bogatcheva G, Washington MK, Coffey RJ. Transformation of polarized epithelial cells by apical mistrafficking of epiregulin. Proc Natl Acad Sci USA. 2013;110(22):8960–8965. doi: 10.1073/pnas.1305508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du P, Kibbe WA, Lin SM. lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013;41(Web Server issue):W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server issue):W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.