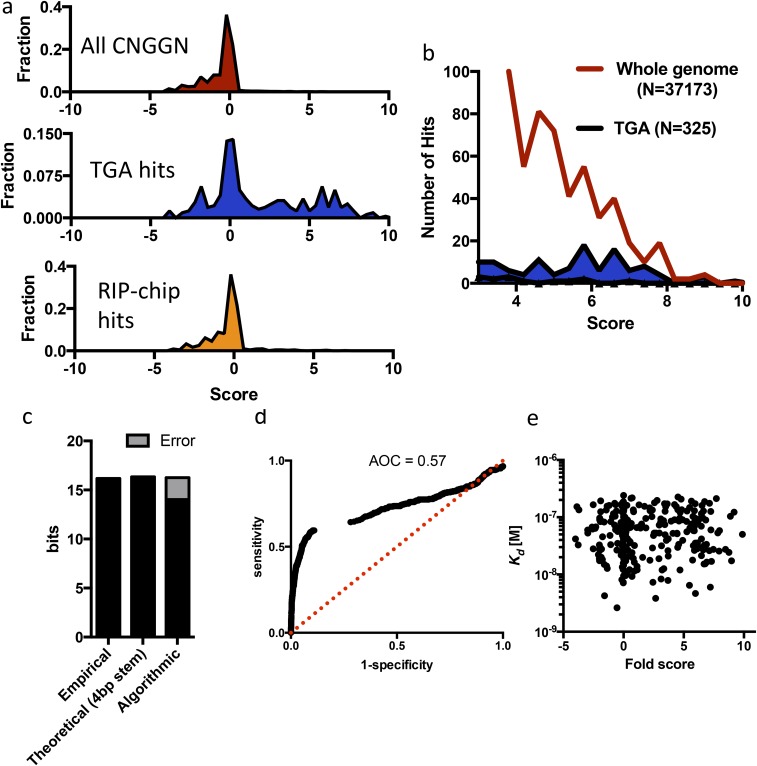
Fig. S5.
(A) NUPACK computational folding output for all CNGGN targets in the genome, targets defined by TGA, and targets defined by RIP-chip (higher score ∝ higher likelihood of CNGGN hairpin). (B) Absolute number of computationally predicted CNGGN hairpins across the entire genome compared with those experimentally identified by TGA. (C) Empirical specificity of Vts1 binding [log2(24 Mb of sequence space/325 targets)] compared with the theoretical informational content of a CNGGN stem loop with 4 bp and the information content captured by the NUPACK folding algorithm. (D) Sensitivity/specificity plot for NUPACK-defined targets compared with TGA-defined targets. (E) Computational fold score does not correlate to Kd measurements.