Significance
Fusion genes resulting from recurrent chromosomal translocations are associated with several tumor types. Cell lines carrying translocations represent important models to study the oncogenic properties of fusion proteins and develop therapeutic strategies. The desmoplastic small round cell tumor is a rare sarcoma characterized by translocation between the EWSR1 and WT1 genes, resulting in expression of the EWSR1–WT1 fusion. A paucity of isogenic cell lines expressing the fusion protein has hindered our understanding of mechanisms associated with tumor development. Here we develop a strategy using CRISPR-Cas9 genome editing to select cells harboring the EWSR1–WT1 translocation with conditional fusion protein expression. This system provides a flexible tool that can be adapted to any translocation or rearrangement in cell lines.
Keywords: CRISPR-Cas9, chromosomal translocation, homology-directed repair, EWSR1–WT1, DSRCT
Abstract
Gene editing techniques have been extensively used to attempt to model recurrent genomic rearrangements found in tumor cells. These methods involve the induction of double-strand breaks at endogenous loci followed by the identification of breakpoint junctions within a population, which typically arise by nonhomologous end joining. The low frequency of these events, however, has hindered the cloning of cells with the desired rearrangement before oncogenic transformation. Here we present a strategy combining CRISPR-Cas9 technology and homology-directed repair to allow for the selection of human mesenchymal stem cells harboring the oncogenic translocation EWSR1–WT1 found in the aggressive desmoplastic small round cell tumor. The expression of the fusion transcript is under the control of the endogenous EWSR1 promoter and, importantly, can be conditionally expressed using Cre recombinase. This method is easily adapted to generate any cancer-relevant rearrangement.
Genomic rearrangements such as deletions, inversions, and translocations are associated with most cancers (1–3). Among these, recurrent chromosomal translocations are implicated in the etiology of several tumor types, including lymphomas, leukemias, and some solid tumors. A translocation can either fuse two genes within intronic sequences to result in expression of a novel fusion protein with oncogenic potential or bring a protooncogene near enhancer or promoter elements, increasing its expression.
The consequences of fusion gene expression due to chromosomal translocation have been widely studied using ectopic expression of the fusion gene products in cell lines or their silencing in tumor cells (4–7). However, both methods present limitations due to nonphysiological levels of fusion gene product expression or incomplete silencing of the fusion gene. Further, the use of patient-derived cell lines to identify compounds that target oncogenic fusion gene products are not ideal due to the presence of tumor-acquired mutations and/or because of prior treatment with therapeutic agents. Thus, to study the role of specific translocations in tumorigenesis and to develop therapies for these malignancies, it is important to have a feasible method to readily induce desired chromosomal translocation in cell culture systems.
DNA double-strand breaks (DSBs) occurring on two chromosomes induce translocations (8–12), a process primarily mediated by nonhomologous end joining (NHEJ), a nonconservative DNA repair pathway that involves the joining of DNA ends without the involvement of extensive homology (1, 13). DSB generation in early model studies was achieved through the introduction of I-SceI endonuclease cleavage sites at two genomic loci, expression of I-SceI, and then selection for the translocation using preintroduced marker fragments (8–10). More recently, the development of programmable nucleases has made it possible to cleave endogenous loci without prior modification (14, 15), greatly simplifying genome editing approaches, including translocations. For example, nucleases that use an array of DNA binding domains of zinc fingers (ZFN) or TAL effectors (TALEN) fused with FokI endonuclease cleavage domains have been used to generate chromosomal translocations in mammalian cells (11, 16, 17). More recently, the discovery of RNA-guided nucleases in bacterial adaptive immunity (CRISPR-Cas9) has facilitated DSB introduction at desired genomic loci (18, 19). Using these various programmable nucleases, a number of cancer translocations have been generated in a variety of human cell lines, including EWSR1–FLI1 (ZFNs, CRISPR-Cas9) (11, 20, 21), NPM1–ALK (TALENs, CRISPR-Cas9) (11, 12), and CD74–ROS1 (CRISPR-Cas9) (22). Oncogenic chromosomal inversions have also been engineered in mouse tissues, leading to tumorigenesis (EML4–ALK) (23).
Compared with repair of a single DSB, repair of two DSBs leading to chromosomal translocation is inefficient, typically occurring at frequencies of ∼10−3 to 10−4, as measured by breakpoint junction analysis in human cells (11, 16). Not surprisingly then, recovery of clones containing translocations is laborious, relying on sib-selection (16), and can fail with primary cells with limited passage number if translocation fusion protein expression is not sufficient for cellular transformation. Further, fusion gene expression occurs concomitantly with the translocation, which may alter growth properties of cells harboring translocations within the background cell population, complicating the analysis of early steps of oncogenic transformation. The inefficient recovery of cell clones that harbor an intended translocation together with constitutive expression of the fusion product before recovery of translocation clones are therefore important problems to overcome. Using a combination of CRISPR-Cas9 technology and homology-directed repair (HDR), we report here an approach to generate and select clones harboring a translocation with conditional fusion protein expression, focusing on the EWSR1–WT1 translocation found in the desmoplastic small round cell tumor (DSRCT) (24–26).
Results and Discussion
Induction of Chromosomal Translocations Using HDR and CRISPR-Cas9.
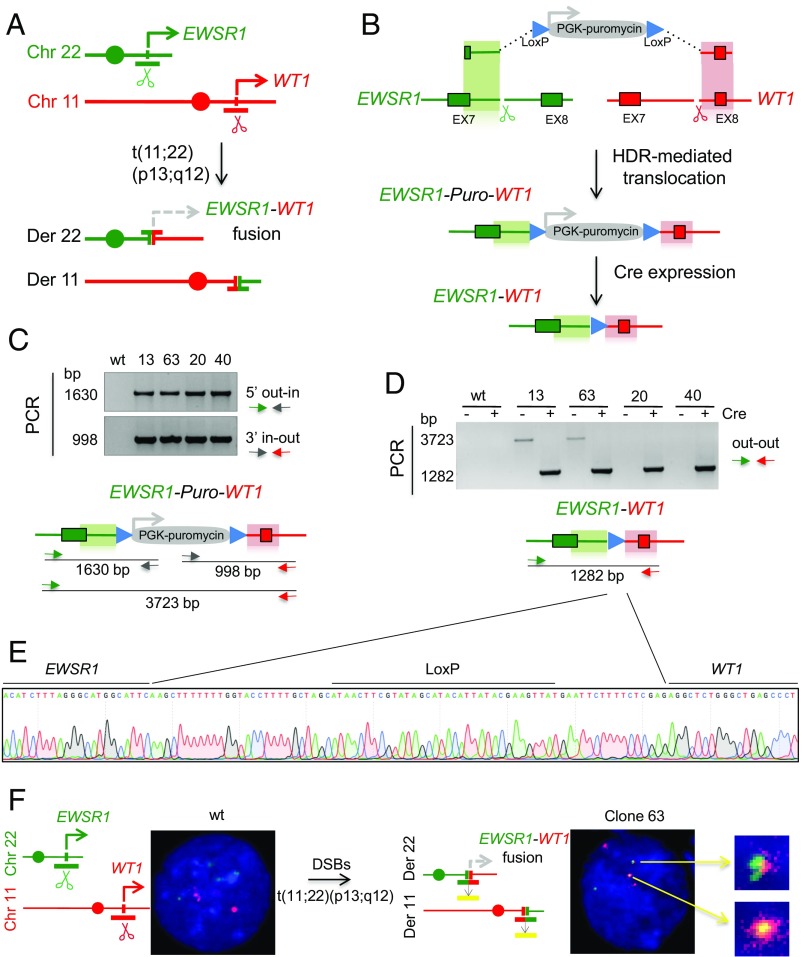
DSRCT is a rare, aggressive neoplasm that derives from mesothelial tissue and is characterized by a t(11;22)(p13;q12) translocation with breakpoints within the EWSR1 and WT1 loci (Fig. 1A) (27). The resulting EWSR1–WT1 chimeric product typically fuses exons 1–7 of EWSR1 to exons 8–10 of WT1, resulting in an in-frame fusion of the transcriptional transactivator domain of EWSR1 to the DNA binding domain of WT1. Because of the scarcity of systems to study DSRCT, we developed a generally applicable approach to isolate clones harboring the EWSR1–WT1 translocation. Further, the strategy was designed for conditional expression of the translocation fusion product through expression of Cre recombinase.
Fig. 1.
Generation of the EWSR1–WT1 chromosomal translocation in immortalized human mesenchymal cells. (A) DSRCT t(11;22)(p13;q12) translocation. Breakpoints within the EWSR1 and WT1 genes on chromosomes 22 and 11, respectively, create the EWSR1–WT1 and the WT1–EWSR1 gene fusions on the derivative chromosomes. (B) HDR targeting strategy to generate chromosomal translocations. A donor plasmid with a puromycin-resistance gene driven by a PGK promoter and flanked by 525-bp homology arms for EWSR1 and WT1 (green and red shading, respectively) undergoes HDR at EWSR1 and WT1 Cas9-induced DSBs generated at the respective gRNA target sites (scissors). The selectable marker is removed by Cre recombinase. (C) Puromycin-resistant clones tested by genomic PCR analysis for correct HDR with the donor plasmid on the 5′ and 3′ homology arms. Primers are located outside the homology in EWSR1 and WT1 and inside the PGK–Puro gene. (D) PCR with primers specific to EWSR1 and WT1 demonstrates the EWSR1–WT1 translocation. The correct 1.3-kb PCR product is seen in all four clones upon Cre expression. Before Cre expression, two clones show the 3.7-kb PCR product consistent with a simple HDR event with the donor DNA (clones 13 and 63). The other two clones (clones 20 and 40) appear to have a more complex HDR event that abrogates PCR before Cre expression. (E) Sanger sequencing of the 1.3-kb PCR product across the EWSR1–WT1 breakpoint junction shows retention of a single LoxP site between the intronic regions of EWSR1 and WT1 after Cre expression for all four clones. (F) Dual-color FISH analysis shows the EWSR1 (green) and WT1 (red) signals for the intact chromosomes and mixed-color signals for the reciprocal EWSR1–WT1 and WT1–EWSR1 derivative chromosomes. Of 32 metaphases analyzed, 28 showed two mixed-colored foci and 4 showed one mixed-colored focus. For the parental cells, no mixed-colored foci were observed in 13 metaphases.
In general terms, the approach makes use of a donor template that contains a selectable marker flanked by short segments (homology arms) derived from the two translocating loci (shaded regions, Fig. 1B). A translocation occurs when a broken end from each chromosomal locus interacts with the cognate homology arm in the donor template to repair the broken end while incorporating the selectable marker. A reciprocal translocation is formed if the other two DNA ends are joined (see below).
Specifically, to generate the EWSR1–WT1 translocation, we used guide RNAs (gRNAs) to result in Cas9-generated DSBs in intron 7 of each of the EWSR1 and WT1 genes (Fig. 1B and Fig. S1A). The donor plasmid for HDR contains EWSR1 and WT1 homology arms of 525 bp each, which flank a drug-resistance marker used to select for integration at the target loci with puromycin. The selectable marker is expressed from its own promoter and can be removed by Cre recombinase.
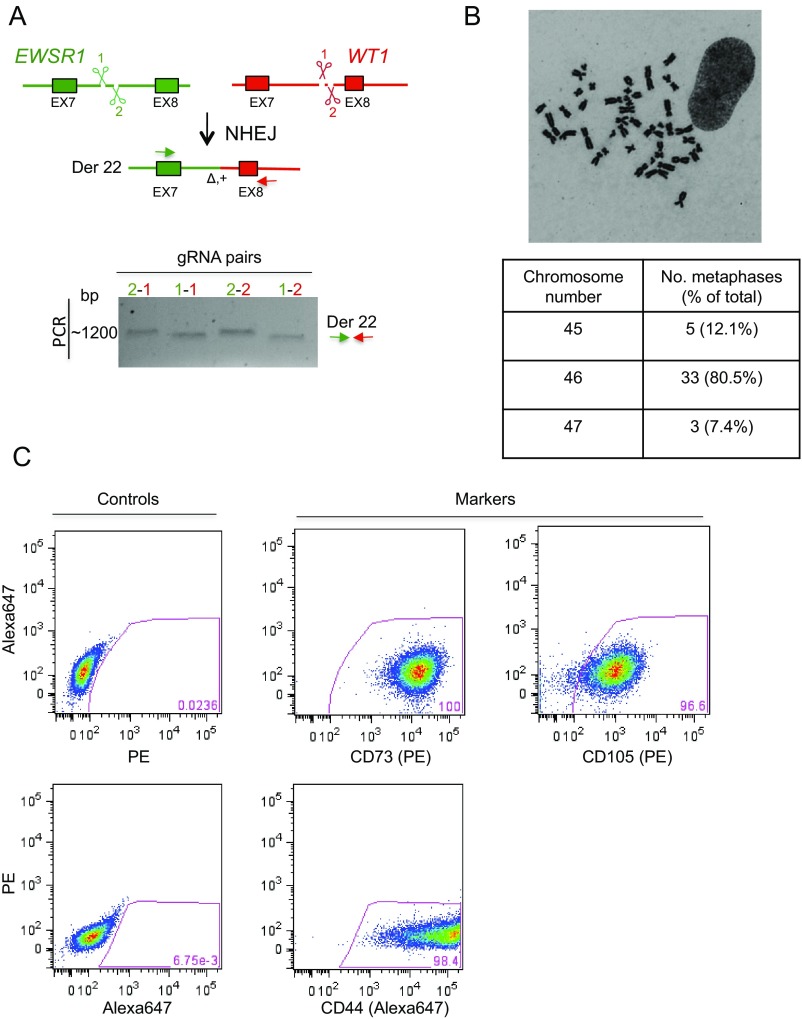
Fig. S1.
Testing of gRNAs and characterization of nonimmortalized mesenchymal stem cells. (A) Induction of DSBs at the EWSR1 and WT1 loci by CRISPR/Cas9 leads to a chromosomal translocation through NHEJ and can be used as a test for gRNA proficiency. For each gene, two gRNAs were designed and all four pairs were tested in the nonimmortalized mesenchymal stem cells and found to be similarly efficient for translocation formation as analyzed by breakpoint junction PCR 48 h after transfection in the absence of the HDR donor plasmid. The same primers were used as for the out–out PCR. The sizes of the bands differ somewhat based on the position of each gRNA target site. (B) Representative metaphase spread and summary table with chromosome number per metaphase of nonimmortalized mesenchymal stem cells. (C) Flow cytometry analysis for various markers characteristic of mesenchymal progenitors: CD44, CD73, and CD105.
We tested this strategy to generate the EWSR1–WT1 translocation in a human telomerase reverse transcriptase (hTERT) immortalized, cyclin-dependent kinase 4 (CDK4)-transformed human mesenchymal stem cell (hMSC) line, given the presumed cell type of origin of DSRCT (28). Cells were transfected with expression vectors for Cas9 and gRNAs for the EWSR1 and WT1 loci and the homologous donor plasmid. At the time of transfection, cells were exposed to a low dose of a DNA-PKcs inhibitor (NU7441) for 2 d to suppress NHEJ and increase HDR levels (29–31).
Puromycin-resistant clones were isolated and PCR analysis was performed on 124 of these clones to detect HDR with the donor plasmid at the EWSR1 and WT1 loci, using one primer specific for each target locus and one for the puromycin-resistance gene (5′ out–in and 3′ in–out PCR, respectively) (Fig. 1C). Four clones showed correct repair of the DSB by HDR at the two loci and were further tested for the fusion of EWSR1 and WT1 using primers outside the homology arms (out–out PCR). After Cre expression, all four clones demonstrated the EWSR1–WT1 fusion separated by the LoxP site (Fig. 1 D and E). Before Cre expression, two of the clones (13 and 63) also gave rise to the out–out PCR product expected for the EWSR1–WT1 fusion containing the intervening puromycin-resistance gene (Fig. 1 C and D). This amplification product was not obtained from the other two clones (20 and 40) upon multiple attempts. Because these two clones show clear evidence of the desired translocation by out–out PCR after Cre expression (see also below), a possible explanation is that the donor plasmid integrated in such a way as to abrogate efficient PCR across the inserted sequences, e.g., a tandem plasmid integration. Thus, screening clones after Cre expression by out–out PCR may identify additional clones harboring translocations.
To formally demonstrate the t(11;22)(p13;q12) translocation, we performed FISH with a dual-fusion probe. Whereas the parental cell line shows discrete green and red signals from EWSR1 and WT1, respectively, overlapping signals indicative of the EWSR1–WT1 were observed with clone 63 (Fig. 1F).
Expression of the EWSR1–WT1 Fusion in Translocation Clones.
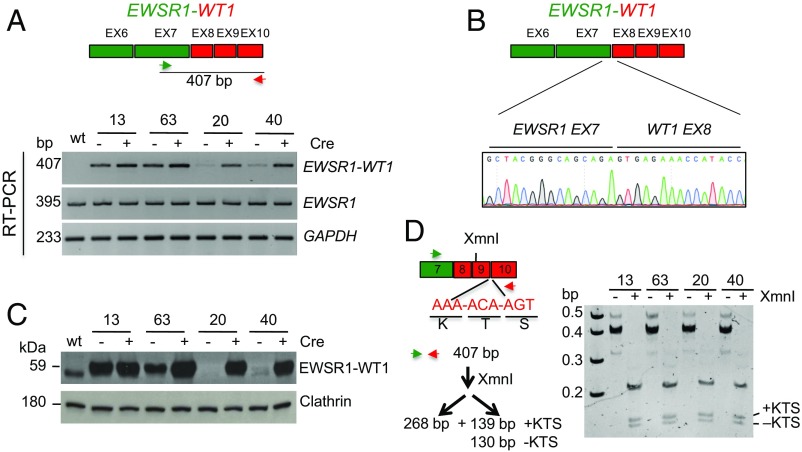
We next performed RT-PCR analysis to detect the EWSR1–WT1 fusion transcript and to determine whether its expression was constitutive or dependent on Cre expression. Using primers to EWSR1 exon 7 and WT1 exon 10, the properly spliced fusion transcript between EWSR1 exon 7 and WT1 exon 8 was observed in clones 13 and 63 both before and after Cre expression, whereas it was only detected at significant levels in clones 20 and 40 after Cre expression (Fig. 2 A and B). Constitutive expression of the EWSR1–WT1 fusion transcript in clones 13 and 63 indicates that the puromycin-resistance gene does not completely interfere with splicing between EWSR1 exon 7 and WT1 exon 8, despite the presence of a polyA site in the sequences inserted in the intron. However, Cre-dependent expression in clones 20 and 40 suggests that the donor integration (e.g., a possible tandem integration) markedly reduces splicing of these exons.
Fig. 2.
Translocation clones express the EWSR1–WT1 fusion product. (A) RT-PCR shows EWSR1–WT1 expression in all clones after Cre expression. Before Cre, expression is also observed in clones 13 and 63, which have undergone a simple HDR event with the donor DNA, indicating that the presence of the PGK–Puro gene in the intron is not sufficient to block splicing of EWSR1 exon 7 to WT1 exon 8. Clones 20 and 40, which have undergone a more complex event, demonstrate conditional expression of EWSR1–WT1. (B) Sanger sequencing of the 407-bp RT-PCR product confirms fusion of EWSR1 exon 7 to WT1 exon 8 in all clones after Cre expression. (C) Western blotting demonstrates EWSR1–WT1 protein expression consistent with the RT-PCR analysis. The antibody is directed against the WT1 C terminus. (D) Both EWSR1–WT1 isoforms are expressed in the translocation clones. The ±KTS isoforms arise from use of alternative WT1 exon 9 splice donors, which affects DNA binding specificity. RT-PCR analysis from post-Cre clones confirms their expression.
To verify that the transcript analysis is reflected in EWSR1–WT1 protein expression, Western blots were performed using an antibody to WT1, which detected the putative 59-kDa fusion protein in all clones after Cre expression (Fig. 2C). Consistent with the RT-PCR results, the EWSR1–WT1 protein was constitutively expressed in clones 13 and 63 but conditionally expressed in clones 20 and 40.
The WT1 portion of the EWSR1–WT1 pre-mRNA is subject to alternative splicing that leads to the retention (or not) of the three amino acids lysine, threonine, and serine, +KTS, between two zinc fingers, which profoundly affects the oncogenic function of the protein (27, 32). Both splice variants are present in the EWSR1–WT1 fusion transcript, with the +KTS variant somewhat more abundant than the −KTS variant (Fig. 2D). Thus, we were able to generate cell lines expressing the EWSR1–WT1 fusion that recapitulates the splice products seen in DSRCT.
Forced Conditional Expression of the EWSR1–WT1 Fusion After Translocation.
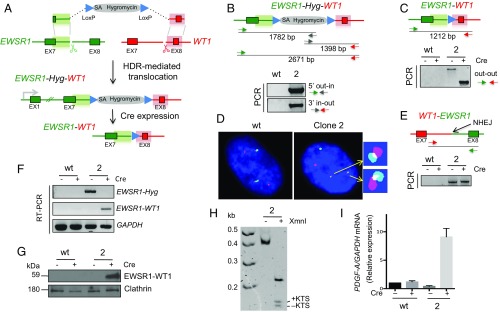
These studies provide a proof of principle for the HDR-based approach for generating and selecting translocations of interest. However, because integration of the transcription unit for the selectable marker between the EWSR1 and WT1 fusion was not sufficient to block fusion protein expression, we modified the approach to incorporate a gene trap, which has been successfully used to knock out genes using Cas9 (e.g., ref. 33). In this strategy, a splice acceptor site is present in the donor plasmid in such a way as to lead to expression of a selectable marker, in this case hygromycin resistance, from the EWSR1 promoter upon HDR with the locus (Fig. 3A). Importantly, the splice acceptor associated with the selectable marker would be predicted to prevent expression of the EWSR1–WT1 fusion transcript after translocation. As with the prior approach (Fig. 1B), the selectable marker is flanked by LoxP sites and so can be deleted by Cre expression, leading to EWSR1–WT1 expression.
Fig. 3.
Gene trap strategy to generate the EWSR1–WT1 translocation in nonimmortalized human mesenchymal stem cells with conditional fusion protein expression. (A) HDR targeting strategy using a gene trap approach. A splice acceptor (SA) upstream of a hygromycin selectable marker in the donor plasmid enables selection of the HDR-mediated t(11;22)(p13;q12) translocation. Removal of the selectable marker by Cre enforces conditional EWSR1–WT1 fusion protein expression. (B) Genomic PCR analysis on both sides of the translocation shows correct HDR with the donor plasmid at the EWSR1 and WT1 loci. Fig. S2A for additional clonal analysis. (C) PCR across the breakpoint junction before and after Cre recombinase expression. A shift in the PCR product size shows loss of the hygromycin selectable marker following Cre expression. (D) Interphase FISH analysis showing the t(11;22) reciprocal translocation, EWSR1–WT1 and WT1–EWSR1, and unrearranged chromosomes 11 and 22. The FISH scheme is the same as that in Fig. 1F. (Fig. S2 B and C for sequence analysis of breakpoint junctions.) (E) NHEJ-mediated reciprocal WT1–EWSR1 fusion in clone 2, which is unchanged after Cre expression. (F) RT-PCR and (G) Western analysis demonstrating conditional EWSR1–WT1 expression. Before Cre expression, only the EWSR1–Hyg transcript is observed, whereas post-Cre, only the EWSR1–WT1 transcript is observed, together with the fusion protein. (H) Polyacrylamide gel electrophoresis of RT-PCR products confirms expression of the ±KTS isoform in the EWSR1–WT1 fusion transcript. (I) Relative fold increase of the EWSR1–WT1 target PDGF-A in clone 2 cells, as demonstrated by quantitative RT-PCR.
We tested this modified approach in genetically unmodified, multipotent human mesenchymal (nonimmortalized) cells derived from human embryonic stem cells (34), which have been previously used to generate nonclonal EWSR1–FLI1 Ewing sarcoma translocations through NHEJ repair (11, 20). Unlike the CDK4-transformed cells (Fig. 1F), these cells maintain a diploid chromosome complement in culture (Fig. S1B). Stable expression of multipotent mesenchymal markers (CD44, CD73, and CD105) is also observed (Fig. S1C) (34). Because these cells are not immortalized, we could address whether EWSR1–WT1 expression is sufficient to transform the cells to overcome growth arrest.
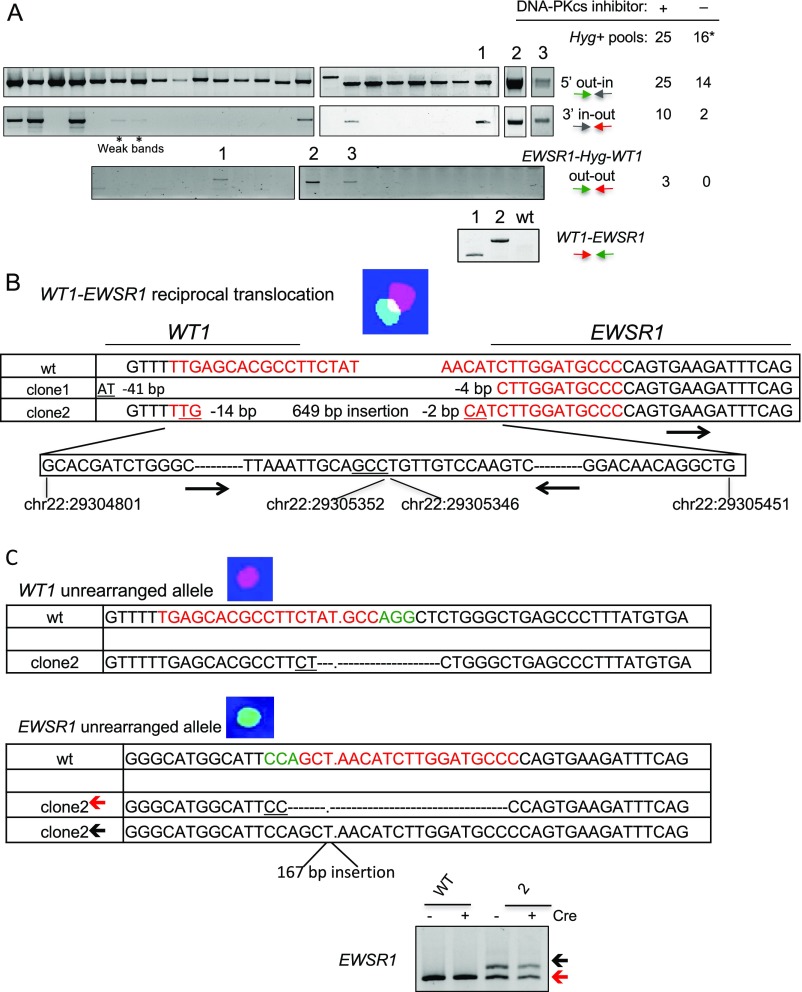
Cells transfected with the new donor plasmid and the Cas9 and gRNA expression vectors were plated at 1,000 cells per well in three 96-well plates and treated with the DNA-PKcs inhibitor for 48 h; 5 d after transfection, cells were exposed to hygromycin for positive selection. Hygromycin-resistant cells grew in 25 wells and were analyzed by PCR for HDR with the donor plasmid at the EWSR1 and WT1 loci (Fig. 3 B and C and Fig. S2A). Cells in all 25 wells had undergone HDR with the donor plasmid at the EWSR1 gene (5′ out–in PCR). Ten of the wells were also positive for HDR with the donor plasmid at the WT1 gene (3′ in–out PCR), three of which were positive for fusion of the EWSR1 and WT1 genes (out–out PCR). Without the DNA-PKcs inhibitor, fewer wells had hygromycin-resistant cells and none of these were positive for the out–out PCR, suggesting that the inhibitor may promote the recovery of clones with a translocation.
Fig. S2.
Analysis of nonimmortalized mesenchymal stem cells after hygromycin selection. (A) Hygromycin-resistant cells were recovered with or without DNA-PKcs inhibitor treatment and analyzed by PCR to investigate correct HDR with the donor plasmid on the 5′ and 3′ sides of the promoterless Hyg gene, the EWSR1–Hyg–WT1 translocation and, for clones 1 and 2, the WT1–EWSR1 reciprocal fusion. Gels are from clones arising after DNA-PKcs inhibitor treatment. The table on the Right indicates the number of Hyg+ clones that test positive by the successive PCR with the tested primer sets. The 16* indicates that two of the 16 clones arising without DNA-PKcs inhibitor treatment did not give rise to the expected out–in PCR; either these clones arose from a random integration that led to Hyg expression or the PCR failed. (B) Sequence analysis of the WT1–EWSR1 fusion. Sequences at the WT1 and EWSR1 breakpoints are indicated. For clone 2, chromosome locations of the inserted EWSR1 sequences are in black with relative orientation indicated by the arrows. Red, gRNA sequences. Underlining shows microhomology. (C) Sequence analysis of the breakpoint junctions for the unrearranged EWSR1 and WT1 alleles in clone 2. Because a single unrearranged EWSR1 locus is observed by FISH, it seems likely that the two EWSR1 breakpoint junctions arose by NHEJ in the next cell division after the translocation. EWSR1 PCR analysis is shown, which demonstrates the two bands. gRNA (red) and protospacer adjacent motif (green) sequences for both genes are indicated in the wild-type sequences.
Cells in two wells grew poorly and in the third (clone 2) grew slowly but could be expanded for further analysis. Upon Cre expression, the direct fusion of EWSR1 and WT1 was confirmed by a shift in the PCR fragment size (Fig. 3C), and sequencing demonstrated retention of the LoxP site at the fusion, as in Fig. 1E.
Cells were tested by FISH before Cre expression to verify the t(11;22)(p13;q12) translocation (Fig. 3D). Whereas the diploid parental cells gave two signals each for chromosomes 11 and 22, clone 2 cells showed only one signal each for intact chromosomes 11 and 22 but additional signals for two derivative chromosomes, indicating a reciprocal translocation.
Whereas the EWSR1–WT1 fusion forms by HDR, the WT1–EWSR1 fusion would be expected to form by NHEJ because the donor plasmid does not have homology to these other two chromosome ends. To determine the nature of the fusion event, PCR was performed across the expected breakpoint junction, giving rise to a fragment that was unchanged in size upon Cre expression (Fig. 3E). Sequencing confirmed the WT1 and EWSR1 fusion (Fig. S2B). The WT1–EWSR1 junction apparently arose by NHEJ with an insertion, as found in other oncogenic translocations (e.g., see ref. 11). The WT1–EWSR1 junction was sequenced from a second clone (clone 1), which also showed an NHEJ event, in this case a simple deletion. We also examined the unrearranged chromosomes in clone 2 to determine whether the gRNA target sites were also cleaved on them; both showed NHEJ junctions at the gRNA sites within the respective EWSR1 and WT1 introns which would have abrogated gRNA binding and subsequent rounds of cleavage by Cas9 (Fig. S2C).
Before Cre expression, RT-PCR showed expression of the EWSR1–Hyg transcript, whereas the EWSR1–WT1 fusion was not detectable (Fig. 3F), indicating the effectiveness of the splice acceptor strategy for impeding fusion gene expression. After Cre exposure and deletion of the selectable marker, expression of the EWSR1–WT1 fusion was apparent both at the mRNA (Fig. 3F) and protein (Fig. 3G) levels, demonstrating that the approach leads to conditional expression. The EWSR1–WT1 fusion product also retained both KTS splice variants (Fig. 3H).
In DSRCT, PDGF-A, a potent fibroblast growth factor, has been reported to be a putative target of EWSR1–WT1 (4). Quantitative RT-PCR (qRT-PCR) confirmed robust induction of the PDGF-A transcript following Cre expression (Fig. 3I). These results indicate that our cell system recapitulates properties of the EWSR1–WT1 fusion product seen in DSRCT. However, EWSR1–WT1 was not sufficient to transform the cells to overcome growth arrest: Whereas both control and clone 2 cells proliferated slowly upon subsequent passages, clone 2 ceased proliferating after Cre expression, suggesting that EWSR1–WT1 expression in nonimmortalized cells is not sufficient for cellular transformation. These results are consistent with previous studies of EWSR1–WT1 expression in mouse fibroblasts and suggest that additional “hits” including p53 mutation may be required (35).
Conclusions
Compared with other systems, our approach presents several advantages for studying the etiology of tumors that involve chromosomal translocations: (i) The fusion gene is expressed under the control of the endogenous promoter, unlike frequently used models in which it is ectopically expressed. (ii) Combining CRISPR-Cas9 technology with HDR allows for the selection of translocations, contrasting with previously used approaches requiring laborious sib-selection. (iii) Incorporating a splice acceptor into the donor DNA for HDR generates translocations with conditional fusion protein expression, which will allow a dissection of the initial steps of transformation.
Here we model the EWSR1–WT1 translocation associated with the rare DSRCT using both immortalized and nonimmortalized human mesenchymal stem cells. The advantage to the former is long-term proliferation of the cells, whereas the latter will allow a dissection of steps for cellular transformation. A similar strategy to that reported here has recently been used to generate the EWSR1–FLI1 fusion in HEK293 cells (36), demonstrating the generality of the approach with a different translocation partner and cell line. The system presented here is expected to be adaptable to model any chromosomal translocation (or other genome rearrangement) and provides a flexible and valuable tool to generate in vitro models to study tumors driven by chromosomal translocations.
Methods
Mammalian Cell Culture and Transfection.
Nonimmortalized human mesenchymal stem/precursor cells [provided by the SKI Stem Cell Research Facility at Memorial Sloan-Kettering (MSK)] were derived from human embryonic stem cells (H1 cell line, XY), and cultivated as described (34). The hTERT-immortalized, CDK4-transformed human mesenchymal stem cell line was obtained from Shinji Kohsaka and Marc Ladanyi, MSK, New York, NY. Briefly, hMSCs (Lonza PT-2501) cultured in mesenchymal stem cell growth medium (MSCGM) (Lonza) were immortalized by serial stable introduction of transgenes encoding hTERT and CDK4 using amphotrophic retrovirus [pCX4–CDK4 (37) and pBabe–hTERT, Addgene; retrovirus packaging: pGP, pE-Ampho, and Takara Bio 6161; and packaging cells: 293T cells, ATCC CRL-3216], selecting in media containing blasticidin (2 mg/mL) and hygromycin (200 μg/mL) to obtain immortalized cells.
Cells were transfected with 2.5 µg of each plasmid (donor plasmid and gRNAs) by Amaxa technology using nucleofector II (program B-016) and treated for 48 h with 0.5 µM DNA-PKcs inhibitor (NU7441), which had no discernible effect on cell viability. The hTERT-immortalized, CDK4-transformed mesenchymal cells were then plated at low density, and colonies from single cells were selected for 10 d in 1 µg/mL puromycin. The nonimmortalized cells were plated in three 96-well plates at a density of 1,000 cells per well, and, after treatment with DNA-PKcs inhibitor, expanded for 5 d in normal media before selection with 50 µg/mL hygromycin for 8 d.
Generation of the Donor for HDR Targeting and gRNAs.
For the generation of the puromycin-based donor plasmid, 525-bp-long homology arms from EWSR1 and WT1 were PCR amplified from genomic DNA from the hTERT-immortalized, CDK4-transformed mesenchymal cells and cloned into MV-PGK-Puro-TK (Transposagen SGK-005) at NotI–HindIII and XhoI–AscI sites (underlined), respectively, using the following primers (36): EWSR1-F-HA 5′-TTAGCGGCCGCCAAACTGGATCCTACAGCCAA-3′, EWSR1-R-HA 5′-CTAAAGCTTGAATGCCATGCCCTAAAGAT-3′, WT1-F-HA 5′-CCACTCGAGAGGCTCTGGGCTGAGCC-3′, WT1-R-HA 5′-TAAGGCGCGCCATGGCTGACTCTCTCATTCATATTC-3′. Note that the PGK-Puro-TK transcription unit has an SV40 polyA site.
The sequence of the EWSR1 homology arm derived from the transformed cells was verified to be identical in the nonimmortalized mesenchymal stem cells. A vector containing a hygromycin-resistance coding sequence with a splice acceptor (SA-2A module) and flanked by LoxP sites (38) was altered to contain the 525-bp EWSR1 homology arm and a 1-bp deletion at the start of the hygromycin-resistance gene to maintain the reading frame with EWSR1. A 520-bp WT1 homology arm was PCR amplified from genomic DNA from the nonimmortalized human mesenchymal stem cells and cloned into SalI–ApaI sites downstream of the second LoxP site: WT1-F-HA2 5′-AAAAAGTCGACCTGGGCTGAGCCCTTTATGT-3′, WT1-R-HA2 5′-AAAAAGGGCCCATGGCTGACTCTCTCATTCA-3′.
gRNAs sequences for EWSR1 and WT1 were cloned into the dual Cas9/gRNA expression vector pSpCas9(BB)-2A-GFP (PX458, Addgene 48138) according to published protocols (39). Oligos for cloning the EWSR1 and WT1 gRNAs are as follows: gRNAEWSR1-1, sense 5′-CACCGAAAAACTCCAAACGTGGCT-3′ and antisense 5′-AAACAGCCACGTTTGGAGTTTTTC-3′; gRNAEWSR1-2 (36), sense 5′-CACCGGGGCATCCAAGATGTTAGC-3′ and antisense 5′-AAACGCTAACATCTTGGATGCCCC-3′;gRNAWT1-1, sense 5′-CACCGTGAGCACGCCTTCTATGCC-3′ and antisense 5′-AAACGGCATAGAAGGCGTGCTCAC-3′; and gRNAWT1-2, sense 5′-CACCGGGCTGAGCCCTTTATGTGA-3′ and antisense 5′-AAACTCACATAAAGGGCTCAGCCC-3′. Underlined sequences represent DNA sequences bound by the gRNAs. All four pairwise combinations of gRNAs were similarly efficient for their ability to generate NHEJ-based translocations (Fig. S1A). gRNA pairs EWSR1-2 and WT1-1 were used to generate the HDR-driven translocations.
PCR Analysis.
Puromycin- and hygromycin-resistant cells were screened for correct HDR with the donor plasmid as previously described (33). The 5′ junction between the donor DNA and EWSR1 was verified by PCR with the following primers: EWSR1ext 5′-TCTCAGCAGAACACCTATGG-3′; Puro-Rev 5′-GGAGGCCTTCCATCTGTTGC-3′ or Hygro-Rev 5′-CCACTATCGGCGAGTACTTC-3′. The 3′ junction between the donor DNA and WT1 was verified by PCR with the following primers:WT1ext 5′-AGGAGGAACATCTCCAGAGA-3′; TK-For 5′-GCGACCTGTACAACGTGTTT-3′; or Hygro-For 5′-GTATCACTGGCAAACTGTGATGG-3′.
The WT1-EWSR1 breakpoint junctions were cloned by PCR with the following primers: WT1intron7 5′-GGATTCTCCTAAGAAGGTGG-3′ and EWSR1exon8 5′-GTTATCAGGGCCACTCATGC-3′. The 5′ and 3′ junction PCRs were performed using the Thermo Scientific Dream Taq Green PCR master mix (Thermo Scientific) under the following conditions: 96 °C for 3 min; 30 cycles of 96 °C 30 s, 60 °C 30 s, 65 °C 2 min, and a final extension step of 65 °C for 5 min.
Translocation was verified by PCR across the breakpoint junction (out–out) using EWSR1ext and WT1ext primers with Jeffreys’ buffer [450 mM Tris HCl pH 8.8, 110 mM (NH4)2SO4, 45 mM MgCl2, 67 mM β-mercaptoethanol, 44 µM EDTA, 10 mM dATP, 10 mM dCTP, 10 mM dGTP, 10 mM dTTP, 1.13 mg/mL BSA, 12.5 mM Tris] with Taq DNA polymerase (ABgene AB-0192) and cloned Pfu DNA polymerase (Agilent Technologies 600254) under the following conditions: 96 °C for 3 min; 30 cycles of 96 °C 30 s, 58 °C 30 s, 65 °C 3.5 min, and a final extension step of 65 °C for 5 min.
The unrearranged EWSR1 allele was PCR amplified using EWSR1ext and EWSR1-Rev (5′-ACATTAATGACTGATAGGG-3′), and the unrearranged WT1 allele was PCR amplified using WT1intron7 and WT1ext. The same conditions were used as for out–out PCR across the breakpoint junction with a 2-min extension time.
Cre Expression.
Puromycin-resistant mesenchymal cells were transfected with a Cre expression plasmid with a CAGGS promoter and mCherry marker (a gift from Federico Gonzalez Grassi, MSK, New York, NY) and plated in a 10-cm plate at low density. After 10 d in regular media, 20 colonies were picked and PCR analysis was performed using primers EWSR1ext and WT1ext to confirm loss of the puromycin-resistant gene. Cells were also grown for 7 d in 1 µg/mL puromycin to confirm sensitivity to puromycin. To remove the hygromycin-resistance gene from the nonimmortalized mesenchymal stem cells, they were infected with a self-deleting lentivirus expressing Cre (40); 72 h postinfection, cells were collected for PCR using the EWSR1ext and WT1ext primers.
RT-PCR.
Total RNA was extracted using RNeasy mini kit (Qiagen) and cDNA was synthesized starting from 100 ng RNA using the SuperScript III First Strand cDNA Syhthesis Kit (Life Technologies). RT-PCR was done using Taq Green PCR master mix with 30 ng cDNA and the fusion product was amplified using the EWSR1-ext and WT1-exon10 primers with the same PCR conditions used to verify correct 5′ and 3′ integration. The unrearranged EWSR1 allele was amplified using primers EWSR1-ex6 and EWSR1-ex8. As reference, GAPDH was amplified with GAPDH-F and GAPDH-R. Conditions for both RT-PCR reactions were 96 °C for 3 min; 30 cycles of 96 °C 30 s, 60 °C 30 s, 65 °C 30 s, and a final extension step of 65 °C for 5 min: WT1-exon10 5′-GACCGGGCAAACTTTTTCTG-3′; EWSR1-ex6 5′-GTAACTACAGTTATCCCCAG-3′; EWSR1-ex8 5′-GTTATCAGGGCCACTCATGC-3′; GAPDH-F 5′-GAGGGGCCATCCACAGTCTTCT-3′; and GAPDH-R 5′-GGAGCCAAAAGGGTCATCATCT-3′.
qRT-PCR was done on Step-One Plus (Applied Biosystems) with Power SYBR Green Master Mix (Thermo Fisher Scientific). Quantification of gene expression was performed with the ΔΔCT method, using GAPDH as a standard. No-template reactions were performed as negative controls.
To characterize the ±KTS variants, the EWSR1–WT1 transcript was amplified and the RT-PCR product was digested with XmnI (NEB) and loaded onto a 4% (wt/vol) polyacrylamide gel.
Western Blotting.
Cells were lysed in TEGN [10 mM Tris pH 8, 1 mM EDTA, 10% (vol/vol) glycerol, 0.5% Nonidet P-40, 400 mM NaCl] with 1 mM DTT in the presence of protease inhibitor (Roche) buffer containing protease inhibitors (Complete, Roche). Lysates were cleared by centrifugation (15,339 x g, 10 min at 4 °C) and 20 µg of protein lysate was separated on a 10% (wt/vol) SDS-polyacrylamide gel. After transfer to a nitrocellulose membrane, blots were blocked in 5% (wt/vol) milk prepared in PBS with 0.1% Tween-20 and probed overnight at 4 °C with anti-WT1 [1:2,000 in 5% (wt/vol) milk, Abcam ab15249] and anti-clathrin [1:10,000 in 5% (wt/vol) milk, BD 610500]. After washing, membranes were incubated with anti-rabbit and anti-mouse horseradish peroxidase-linked secondary antibody [1:10,000 in 5% (wt/vol) milk] for 1 h. Proteins were visualized with Western Lightning Plus-ECL Enhanced Chemiluminescence (Perkin-Elmer).
Chromosome Analysis.
FISH was performed on metaphases from hTERT-immortalized, CDK4-transformed human mesenchymal stem cell (wild type and EWSR1–WT1 clones) using a FITC-labeled probe for EWSR1 (LPS007, Cytocell) and a Cy5-dUTP-labeled (PerkinElmer, NEL579001EA) BAC probe for WT1 (RP1-74J1, CHORI). Metaphase spreading, probe labeling, hybridization, washing, and fluorescence detection were performed according to standard procedures. Images were taken with the Zeiss Axio Observer Z1 system. Mixed-colored foci represent the EWSR1–WT1 fusion. Nonimmortalized mesenchymal stem cells were fixed in Carnoy’s fixative and hybridized with green EWSR1 probe (Hg19 chr22: 29,380,432–29,980,251) and red WT1 probe (Hg19 chr11: 32,133,248–32,733,248), according to the protocol suggested by Agilent Technologies.
For metaphase spreads, 1 million cells were treated with colcemid (0.3 mg/mL final) for 40 min, trypsinized, and incubated for 10 min in KCl (0.075 M) at 37 °C before being fixed with 3 vol methanol:1 vol acetic acid. Cells were resuspended in 0.5 mL fixative solution and 10 µL were spotted on slides before staining with Giemsa/Sorensen buffer for 15 min.
Flow Cytometry Analysis.
Cells were dissociated as a single cell suspension and resuspended at 1 million cells per 100 µL in 1× PBS, 0.1% BSA. Conjugated antibodies were added at indicated dilutions and cells were incubated for 15 min in the dark before washing three times with 1× PBS 0.1% BSA. Cells were finally resuspended in 300 µL 1× PBS and analyzed by BD FACS ArialII. Antibodies were as follows: CD73 (mouse anti-human CD73-PE, BD Pharmingen 550257), CD105 (mouse anti-human CD105-PE, Serotec MCA 1557PE), and CD44 (rat anti-mouse/human CD44-AF 647, Biolegend).
Acknowledgments
We thank William Gerald, who first described DSRCT, and Beth Elliott for early interest and efforts on the project; Marc Ladanyi and Lee Spraggon for initial discussions on translocation approaches and materials; Lu Wang, Elodie Pronier, and Enrico Velardi for technical assistance and reagents; and Travis White for careful reading of the manuscript. Memorial Sloan Kettering research is supported by NIH/National Cancer Center Support Grant P30 CA008748. This work was supported in part by an Alex’s Lemonade Stand Innovation Award, NIH R01CA185660 and R35GM118175 (to M.J.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700622114/-/DCSupplemental.
References
- 1.Mani RS, Chinnaiyan AM. Triggers for genomic rearrangements: Insights into genomic, cellular and environmental influences. Nat Rev Genet. 2010;11(12):819–829. doi: 10.1038/nrg2883. [DOI] [PubMed] [Google Scholar]
- 2.Elliott B, Jasin M. Double-strand breaks and translocations in cancer. Cell Mol Life Sci. 2002;59(2):373–385. doi: 10.1007/s00018-002-8429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieber MR. Mechanisms of human lymphoid chromosomal translocations. Nat Rev Cancer. 2016;16(6):387–398. doi: 10.1038/nrc.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SB, et al. The EWS-WT1 translocation product induces PDGFA in desmoplastic small round-cell tumour. Nat Genet. 1997;17(3):309–313. doi: 10.1038/ng1197-309. [DOI] [PubMed] [Google Scholar]
- 5.Braunreiter CL, Hancock JD, Coffin CM, Boucher KM, Lessnick SL. Expression of EWS-ETS fusions in NIH3T3 cells reveals significant differences to Ewing’s sarcoma. Cell Cycle. 2006;5(23):2753–2759. doi: 10.4161/cc.5.23.3505. [DOI] [PubMed] [Google Scholar]
- 6.Werner H, et al. A novel EWS-WT1 gene fusion product in desmoplastic small round cell tumor is a potent transactivator of the insulin-like growth factor-I receptor (IGF-IR) gene. Cancer Lett. 2007;247(1):84–90. doi: 10.1016/j.canlet.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Lessnick SL, Ladanyi M. Molecular pathogenesis of Ewing sarcoma: New therapeutic and transcriptional targets. Annu Rev Pathol. 2012;7:145–159. doi: 10.1146/annurev-pathol-011110-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405(6787):697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 9.Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell. 2005;17(6):885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 10.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: Differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107(2):777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piganeau M, et al. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013;23(7):1182–1193. doi: 10.1101/gr.147314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghezraoui H, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55(6):829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res. 2013;2(3):130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 15.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunet E, et al. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci USA. 2009;106(26):10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simsek D, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7(6):e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renouf B, Piganeau M, Ghezraoui H, Jasin M, Brunet E. Creating cancer translocations in human cells using Cas9 DSBs and nCas9 paired nicks. Methods Enzymol. 2014;546:251–271. doi: 10.1016/B978-0-12-801185-0.00012-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres R, et al. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun. 2014;5:3964. doi: 10.1038/ncomms4964. [DOI] [PubMed] [Google Scholar]
- 22.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddalo D, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerald WL, Rosai J. Case 2. Desmoplastic small cell tumor with divergent differentiation. Pediatr Pathol. 1989;9(2):177–183. doi: 10.3109/15513818909022347. [DOI] [PubMed] [Google Scholar]
- 25.Gerald WL, Rosai J. Desmoplastic small cell tumor with multi-phenotypic differentiation. Zentralbl Pathol. 1993;139(2):141–151. [PubMed] [Google Scholar]
- 26.Gerald WL, Haber DA. The EWS-WT1 gene fusion in desmoplastic small round cell tumor. Semin Cancer Biol. 2005;15(3):197–205. doi: 10.1016/j.semcancer.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Gerald WL, Rosai J, Ladanyi M. Characterization of the genomic breakpoint and chimeric transcripts in the EWS-WT1 gene fusion of desmoplastic small round cell tumor. Proc Natl Acad Sci USA. 1995;92(4):1028–1032. doi: 10.1073/pnas.92.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerald WL, et al. Intra-abdominal desmoplastic small round-cell tumor. Report of 19 cases of a distinctive type of high-grade polyphenotypic malignancy affecting young individuals. Am J Surg Pathol. 1991;15(6):499–513. [PubMed] [Google Scholar]
- 29.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15(24):3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen C, Kurimasa A, Brenneman MA, Chen DJ, Nickoloff JA. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc Natl Acad Sci USA. 2002;99(6):3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vriend LE, et al. Distinct genetic control of homologous recombination repair of Cas9-induced double-strand breaks, nicks and paired nicks. Nucleic Acids Res. 2016;44(11):5204–5217. doi: 10.1093/nar/gkw179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haber DA, et al. Alternative splicing and genomic structure of the Wilms tumor gene WT1. Proc Natl Acad Sci USA. 1991;88(21):9618–9622. doi: 10.1073/pnas.88.21.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Vanoli F, LaRocque JR, Krawczyk PM, Jasin M. Biallelic targeting of expressed genes in mouse embryonic stem cells using the Cas9 system. Methods. 2014;69(2):171–178. doi: 10.1016/j.ymeth.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barberi T, Willis LM, Socci ND, Studer L. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med. 2005;2(6):e161. doi: 10.1371/journal.pmed.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bandopadhayay P, et al. The oncogenic properties of EWS/WT1 of desmoplastic small round cell tumors are unmasked by loss of p53 in murine embryonic fibroblasts. BMC Cancer. 2013;13:585. doi: 10.1186/1471-2407-13-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spraggon L, et al. Generation of conditional oncogenic chromosomal translocations using CRISPR-Cas9 genomic editing and homology-directed repair. J Pathol. 2017 doi: 10.1002/path.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasai K, et al. Oncogene-mediated human lung epithelial cell transformation produces adenocarcinoma phenotypes in vivo. Cancer Res. 2011;71(7):2541–2549. doi: 10.1158/0008-5472.CAN-10-2221. [DOI] [PubMed] [Google Scholar]
- 38.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfeifer A, Brandon EP, Kootstra N, Gage FH, Verma IM. Delivery of the Cre recombinase by a self-deleting lentiviral vector: Efficient gene targeting in vivo. Proc Natl Acad Sci USA. 2001;98(20):11450–11455. doi: 10.1073/pnas.201415498. [DOI] [PMC free article] [PubMed] [Google Scholar]