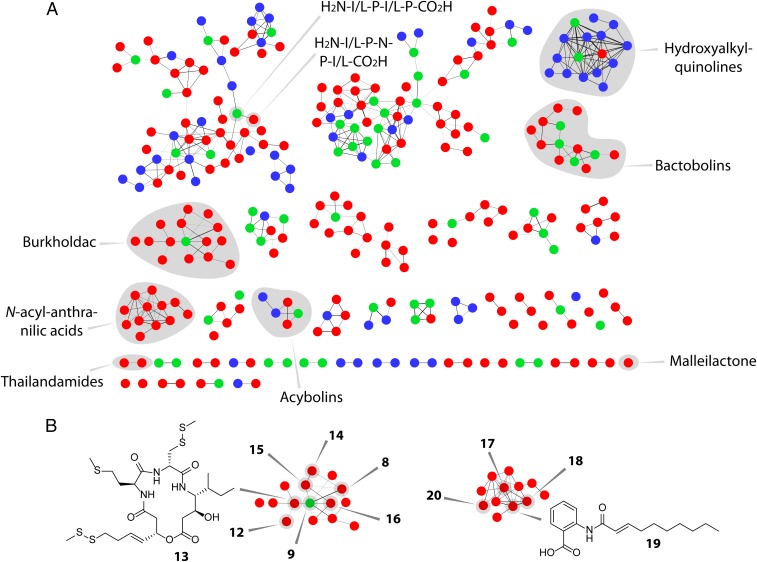
Fig. 4.
MS/MS network analysis of the secondary metabolomes of WT and ΔscmR B. thailandensis E264. (A) Each node corresponds to a unique secondary metabolite. The thickness of a line connecting two nodes is an indicator of structural similarity. Compounds observed only in ΔscmR (red nodes), only in WT (blue nodes), or in both cultures (green nodes) are shown. Molecular families that were identified are shaded in gray and labeled. A large molecular family (top left) appears to consist of peptide fragments; the sequences of two such peptides were identified by tandem HR-MS. The acybolin family contains two previously identified variants (red nodes); other variants within this family were not detected before and have not yet been verified as acybolin relatives. (B) Compounds produced in ΔscmR. Several burkholdac analogs were identified in ΔscmR. The variants identified are marked in the burkholdac molecular cluster (8, 9, and 12–16). Similarly, structures of N-acyl-anthranilic acids are indicated in the corresponding molecular family (17–20; Fig. 1).