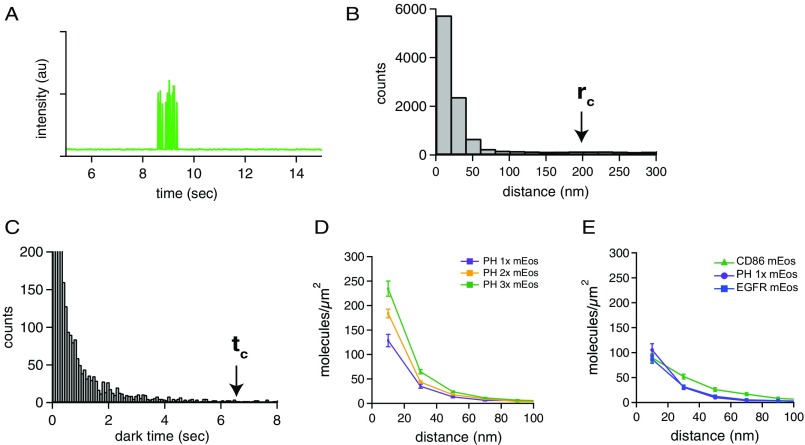
Fig. S2.
Threshold determination for blink correction and PH calibration. (A) An example of a fluorescence time trace of a single mEos-tagged receptor photoactivated with 405-nm light and imaged with 561-nm excitation light. The fluorophore turns on, blinks several times, and then turns off (photo bleaches) permanently. (B) Pairwise distance histogram of raw (not blink-corrected) STORM localizations of a presumed monomeric receptor (PH-1×). The cutoff radius (rc) used for the subsequent blink corrections was set at the 99th percentile of this histogram, around 200 nm. (C) Histogram of the dark times of a sparse, monomeric sample of mEos-tagged PH-1×. Dark times were determined by applying the blink correction to the STORM data, using a very large cutoff time, and then determining the 99th percentile of these dark times to set a threshold for the blink correction of all data. The cutoff time (tc) was determined to be around 6.4 s (400 frames). (D) One, two, or three copies of mEos were fused to the PH domain, a membrane-localized and presumed monomeric protein. These constructs were expressed in NR6 cells and imaged by STORM, and the blink-corrected pairwise distance histograms were calculated. Although the presumed monomeric curve is not flat, as was expected, progressive increases in peak amplitude were observed for the doubly and triply tagged proteins. This basal peak in the pairwise distance histogram could be caused by clustering of the PH domain proteins in the plasma membrane of mammalian cells, by membrane inhomogeneity, or by rare long-lived dark states of the chromophore. Therefore only relative changes in the pairwise distance histogram were used for all analysis. (E) Pairwise distance histogram for mEos-tagged CD86 and mEos-tagged PH-1× constructs plotted on the same graph as mEos–EGFR. All constructs were stably expressed in NR6 cell lines.