Abstract
Purpose
Sixteen different mutations in the guanylate cyclase activator 1A gene (GUCA1A), have been previously identified to cause autosomal dominant cone dystrophy (adCOD), cone–rod dystrophy (adCORD), macular dystrophy (adMD), and in an isolated patient, retinitis pigmentosa (RP). The purpose of this study is to report on two novel mutations and the patients’ clinical features.
Methods
Clinical investigations included visual acuity and visual field testing, fundus examination, high-resolution spectral-domain optical coherence tomography (OCT), fundus autofluorescence imaging, and full-field and multifocal electroretinogram (ERG) recordings. GUCA1A was screened by Sanger sequencing in a cohort of 12 French families with adCOD, adCORD, and adMD.
Results
We found two novel GUCA1A mutations—one amino acid deletion, c.302_304delTAG (p.Val101del), and one missense mutation, c.444T>A (p.Asp148Glu)—each of which was found in one family. The p.Asp148Glu mutation affected one of the Ca2+-binding amino acids of the EF4 hand, while the p.Val101del mutation resulted in the in-frame deletion of Valine-101, localized between two Ca2+-binding aspartic acid residues at positions 100 and 102 of the EF3 hand. Both families complained of visual acuity loss worsening with age. However, the p.Asp148Glu mutation was present in one family with adCOD involving abnormal cone function and an absence of macular atrophy, whereas p.Val101del mutation was encountered in another family with adMD without a generalized cone defect.
Conclusions
The two novel mutations described in this study are associated with distinct phenotypes, MD for p.Val101del and COD for p.Asp148Glu, with no intrafamilial phenotypic heterogeneity.
Introduction
Cone and cone–rod dystrophies (CODs and CORDs, respectively) are rare, genetically heterogeneous diseases inherited as a dominant, recessive, or X-linked trait. COD and CORD are rare hereditary retinal dystrophies with an estimated prevalence of 1 in 40,000 individuals [1]. They are primarily characterized by progressive loss of cone photoreceptor function; for CORD, this is followed by the secondary, or sometimes concomitant, loss of rod photoreceptor function [1]. Typical symptoms of COD and CORD include photophobia, reduced central visual acuity, and loss of color vision [1,2]. Funduscopy has revealed a range of macular phenotypes, from normal or mild changes to severe macular atrophy [3]. The preserved peripheral vision distinguishes COD from CORD.
Ten disease-causing genes (AIPL1, CRX, GUCA1A, GUCY2D, PITPNM3, PROM1, PRPH2, RIMS1, SEMA4A, and UNC119) and four loci (CORD4, CORD12, CORD17, and RCD1) are currently known in autosomal dominant (ad) COD and CORD (RetNet). Among these genes, GUCA1A and GUCY2D encode guanylate cyclase-activating protein 1 (GCAP1) and retinal guanylate cyclase 1 (GC1, also called RetGC1), respectively, which are involved in the synthesis of cGMP from GTP, an important step in the recovery of photoreceptors after light stimulation and cGMP hydrolysis [4,5].
Following light stimulation of the photoreceptors, rhodopsin or cone photopigments activate their downstream target, transducin, which in turn binds the effector enzyme PDE6, resulting in the hydrolysis of intracellular cGMP; thus, the concentration of cGMP is lowered and a closure of cGMP-gated channels, a decrease in the concentration of free Ca2+, the hyperpolarization of the photoreceptor outer segment, and a neural response are produced [6-8]. The GCAP proteins, activated by low Ca2+ concentration, then stimulate the photoreceptor guanylate cyclases, which convert GTP to cGMP. The pool of cGMP is thus replenished and the dark-adapted state is restored [9-11].
GUCY2D and GUCY2F encode the two types of membrane-bound guanylate cyclases expressed in the photoreceptors, namely GC1 and GC2, respectively [12-14]. Both guanylate cyclases are expressed in rods and cones, but GC1 is predominantly expressed in cones [15-18]. Three isoforms of GCAP proteins (GCAP1–3), encoded by GUCA1A, GUCA1B and GUCA1C, respectively, stimulate GC1 or GC2 in low [Ca2+]free [19-21]. GCAP1 is present in higher concentrations in cone than in rod outer segments [20,22]. GCAP2 is localized in rods [19], cone inner segments, somata, synaptic terminals [23], and inner retinal neurons [22,24]. GCAP3 is expressed exclusively in cones [21]. Thus, with the predominant expression of GC1 and GCAP1 in cones, mutations in GUCY2D and GUCA1A result in more severe degeneration in cones than in rods.
To date, 16 different mutations in GUCA1A have been reported in the literature to cause adCOD, adCORD, ad macular dystrophy (adMD), and in an isolated patient, retinitis pigmentosa (RP; Table 1). In this study, the GUCA1A gene was screened for mutations in 12 probands with adCOD, adCORD, and adMD from unrelated French families (Appendix 1). We report on two novel mutations and clinical data associated with the two families with GUCA1A mutations. We found that one family had MD without a generalized cone defect, while the other had typical COD.
Table 1. Summary of GUCA1A gene known and novel mutations.
| Nucleotide change | Exon | Protein change | Region | ExAC | PPhen2 HDIV | PPhen2 HVAR | SIFT | Prov | Mut. taster | Clin. | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| c.149C>T |
3 |
p.Pro50Leu |
EF1-EF2 link |
121/121,374 |
BEN |
BEN |
TOL |
DEL |
DIS |
COD, CORD |
[29] |
| c.250C>T |
4 |
p.Leu84Phe |
EF2 helix F |
0 |
PRO |
PRO |
DAM |
DEL |
DIS |
COD, CORD, MD |
[36] |
| c.265G>A |
4 |
p.Glu89Lys |
EF2-EF3 link |
0 |
POS |
BEN |
DAM |
DEL |
DIS |
COD, MD |
[27] |
| c.296A>G |
4 |
p.Tyr99Cys |
EF3 helix E |
0 |
PRO |
PRO |
DAM |
DEL |
DIS |
COD, CORD, MD |
[33,37] |
| c.299A>G |
4 |
p.Asp100Gly |
EF3 loop, Ca2+ binding |
0 |
PRO |
PRO |
DAM |
DEL |
DIS |
CORD, MD |
[38] |
| c.300T>A |
4 |
p.Asp100Glu |
EF3 loop, Ca2+ binding |
0 |
PRO |
PRO |
DAM |
DEL |
DIS |
COD |
[27] |
| c.302_304delTAG |
4 |
p.Val101del |
EF3 loop |
0 |
N.A. |
N.A. |
N.A. |
DEL |
DIS |
MD |
Present study |
| c.304G>C |
4 |
p.Asp102His |
EF3 loop, Ca2+ binding |
0 |
PRO |
PRO |
DAM |
DEL |
DIS |
CORD |
[41] |
| c.312C>A |
4 |
p.Asn104Lys |
EF3 loop, Ca2+ binding |
0 |
PRO |
POS |
DAM |
DEL |
DIS |
COD |
[42] |
| c.320T>C |
4 |
p.Ile107Thr |
EF3 loop |
0 |
PRO |
PRO |
DAM |
DEL |
DIS |
COD, CORD, MD |
[36] |
| c.341C>T |
4 |
p.Thr114Ile |
EF3 helix F |
5/121,228 |
BEN |
BEN |
TOL |
NEU |
POL |
Atypical RP |
[30] |
| c.428delTinsACAC |
5 |
p.Ile143delinsAsnThr |
EF4 helix E |
0 |
N.A. |
N.A. |
N.A. |
DEL |
POL |
COD |
[30] |
| c.444T>A |
5 |
p.Asp148Glu |
EF4 loop |
0 |
POS |
POS |
TOL |
DEL |
DIS |
COD |
Present study |
| c.451C>T |
6 |
p.Leu151Phe |
EF4 loop |
0 |
PRO |
PRO |
DAM |
DEL |
DIS |
COD |
[27,34] |
| c.464A>G |
6 |
p.Glu155Gly |
EF4 loop, Ca2+ binding |
0 |
PRO |
PRO |
DAM |
DEL |
DIS |
COD |
[35] |
| c.464A>C |
6 |
p.Glu155Ala |
EF4 loop, Ca2+ binding |
0 |
PRO |
PRO |
DAM |
DEL |
DIS |
CORD |
[43] |
| c.476G>T |
6 |
p.Gly159Val |
EF4 helix F |
0 |
PRO |
PRO |
DAM |
DEL |
DIS |
COD, CORD |
[27] |
| c.526C>T | 6 | p.Leu176Phe | C-ter | 0 | PRO | PRO | TOL | NEU | DIS | MD | [39] |
EF, helix E - loop - helix F; Ca2+-binding domain; C-ter, carboxy terminal; ExAC, Exome Aggregation Consortium; COD, cone dystrophy; CORD, cone-rod dystrophy; MD, macular dystrophy; PPhen2, PolyPhen2, POS for possibly damaging, PRO for probably damaging, BEN for benign; SIFT, TOL for tolerated, DAM for damaging Prov; PROVEAN (Prediction with cutoff= -2.5), DEL for deleterious, NEU for neutral; Mut. taster, Mutation taster, POL for Polymorphism and DIS for Disease Causing; N.A., Not applicable; Clin, clinical diagnosis; Ref, reference.s
Methods
Patients
Twelve index patients were included in the study. Informed and written consent was obtained for all patients participating in the study. The study (#2008-A01238–47) received authorization from the Sud méditerranée IV ethical board committee (#08 10 05, 04/11/2008), was approved by the French regulation agency for medication (AFSSAPS #B81319–70), and is registered at Clinical Trials (#NCT01235624). The investigators followed the tenets of the Declaration of Helsinki.
Clinical investigations
Patients underwent a standard ophthalmologic examination (refractometry, visual acuity, slit-lamp examination, applanation tonometry, funduscopy). Kinetic visual fields were determined with a Goldmann perimeter with targets V4e, III4e, and I4e. Optical coherence tomography (OCT) measurement of the macula was performed using an OCT-3 system (Stratus model 3000, Carl Zeiss Meditec, Dublin, CA) or with a spectral domain OCT (Spectralis, Heidelberg, Germany) with software version 3.0. Autofluorescence measurements were obtained with the HRA2 Heidelberg retinal confocal angiograph (Heidelberg Engineering, Dossenheim, Germany), and fundus pictures were taken. Full-fields ERG was recorded using a ganzfeld apparatus (Metrovision, Pérenchies, France) with a bipolar contact lens electrode on maximally dilated pupils according to the International Society for Clinical Electrophysiology of Vision (ISCEV) protocol. Multifocal ERGs were recorded by pattern stimulations of the central 20° with small corneal electrodes using the Reti-port system (Roland Consult, Brandenburg, Germany).
Mutation screening
Genomic DNA was isolated from 10 ml of peripheral blood leucocytes using a standard salting-out procedure [25]. Coding exons and adjacent intronic sequences of GUCA1A gene (NM_000409.3; primer pairs and PCR conditions in Appendix 2) were sequenced with an Applied Biosystems 3130xl genetic analyzer (Applied Biosystems, Foster City, CA) using a BigDye Terminator cycle sequencing ready reaction kit V3.1 (Applied Biosystems) following the manufacturer’s instructions. Sequence analysis and mutation identification were performed using the Collection and Sequence Analysis software package (Applied Biosystems). Sorting Intolerant from Tolerant (SIFT), Polymorphism Phenotyping 2 (PolyPhen2), Protein Variation Effect Analyzer (PROVEAN), and MutationTaster were used to predict the possible effects of missense variants.
Results
Identification of novel GUCA1A mutations
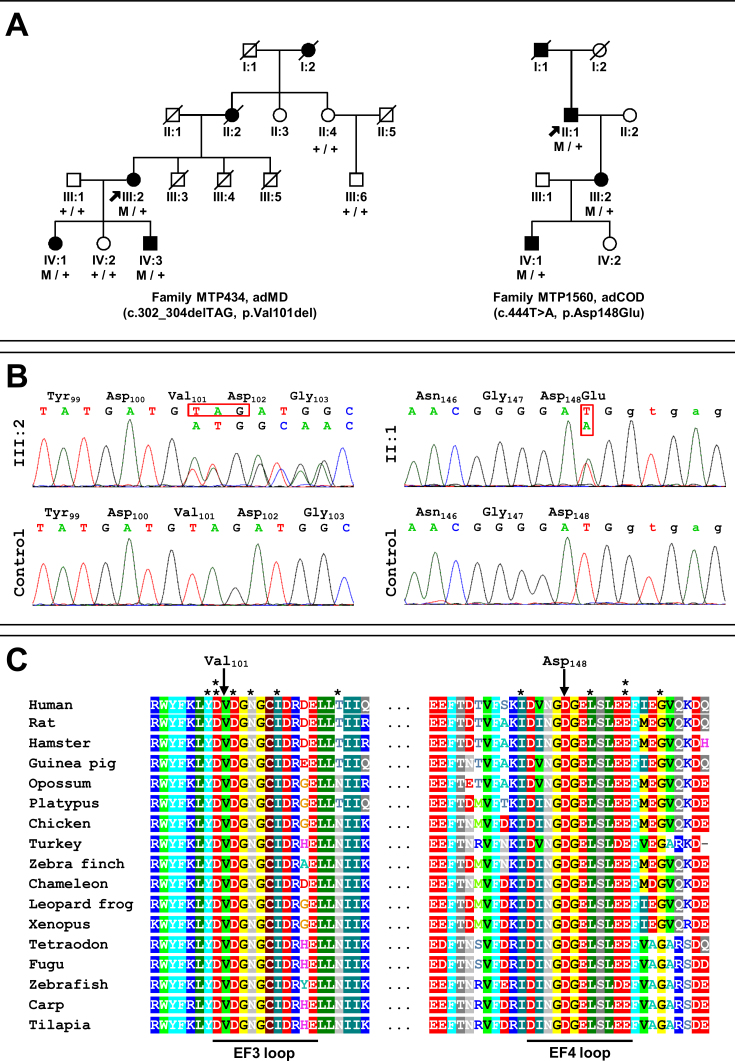
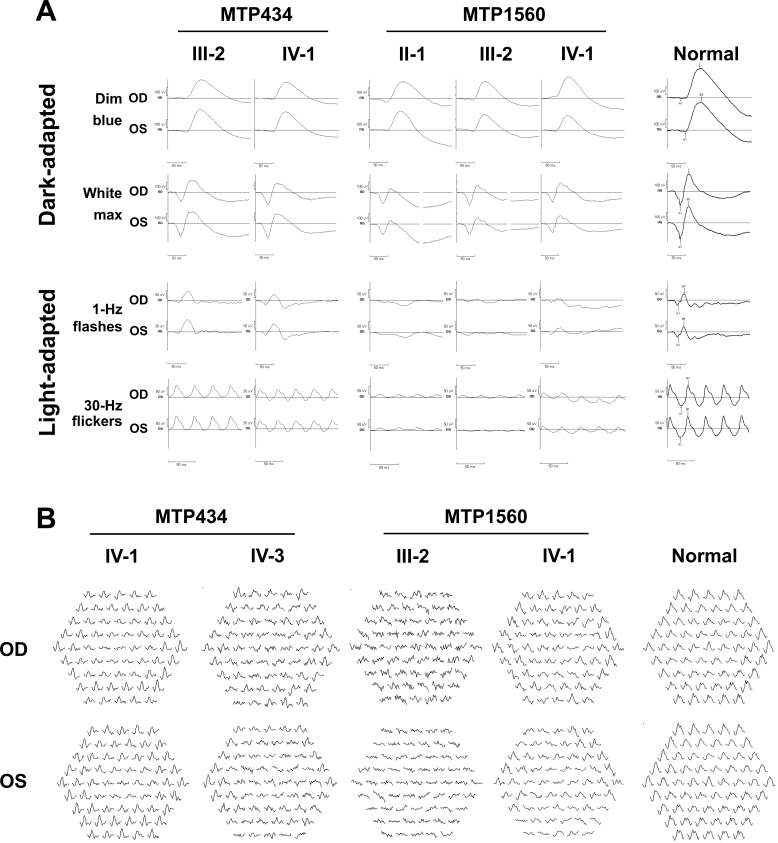
A cohort of 12 French families (Appendix 1) with adCOD, adCORD), and adMD was screened for the six exons of GUCA1A (NM_000409.3), including the noncoding exons 1 and 2. We identified two novel mutations (16.7%) in the families, namely MTP434 and MTP1560 (Figure 1A). We found c.302_304delTAG (p.Val101del) in MTP434 and c.444T>A (p.Asp148Glu) in MTP1560 (Figure 1B). Both mutations cosegregated with the disease phenotype in available family members (Figure 1A). They were not found in public human genetic databases, including Ensembl, HapMap, the 1000 Genomes project, dbSNP, Exome Variant Server (EVS; release ESP6500), or Exome Aggregation Consortium (ExAC). The substitution p.Asp148Glu was predicted to be damaging by the PolyPhen2, PROVEAN, and MutationTaster programs, but not by SIFT (Table 1). The c.302_304delTAG mutation led to the in-frame deletion of valine at position 101 (p.Val101del). Both affected amino acids, valine 101 and aspartic acid 148, were conserved in 16 GCAP1 orthologs (Figure 1C). Both mutations were localized in the 12-amino-acid loop of one of the EF-hand domains of the protein. The mutation p.Asp148Glu affected one of the Ca2+-binding amino acids of the EF4 hand, and the mutation p.Val101del resulted in the in-frame deletion of one amino acid, valine 101, localized between two Ca2+-binding aspartic acid residues at positions 100 and 102 of the EF3 hand (Figure 2).
Figure 1.
Pedigrees of families with autosomal dominant cone dystrophy (adCOD) or macular dystrophy (adMD) and with the novel mutations in GUCA1A identified in this study. A: Filled symbols indicate affected family members; squares: males; circles: females; arrows: index patients. Family MTP434 with the c.302_304delTAG (p.Val101del) mutation. Family MTP1560 with the c.444T>A (p.Asp148Glu) mutation. B: Electropherograms show the normal control and affected sequences (index individuals III:2 and II:1) surrounding the c.302_304delTAG and c.444T>A mutations. C: Multiple amino acid sequence alignment of GCAP1 for a region surrounding the novel p.Val101del and the p.Asp148Glu mutations. The site of the mutation is indicated by an arrow. The 12-amino-acid loop of the EF3- and EF4-hand domains of the protein are underlined and the 12 referenced mutations within or flanking the loops are indicated with a star (*; Y99C, D100G, D100E, D102H, N104K, I107T, T114I, I143NT, L151F, E155G, E155A, G159V).
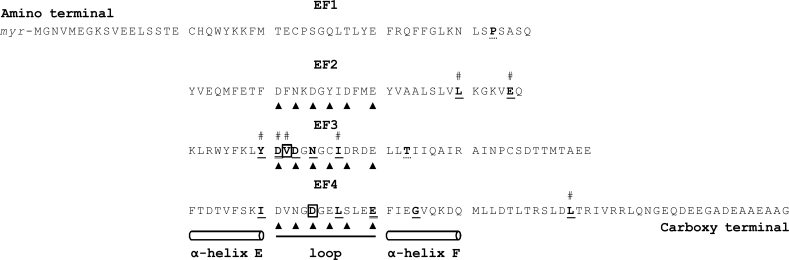
Figure 2.
Summary of the referenced and the novel mutations on the GCAP1 primary structure. Schematic representation of the guanylate cyclase-activating protein 1 (GCAP1) showing the location of the two novel mutations presented in this study (in bold and framed) and the known mutations (in bold and underlined; in bold and double underlined when two different mutations are localized on the same amino acid; in dashed underline for unlikely mutations). The EF-hand domains consist of an alpha-helix (E), a 12-amino-acid loop, and a second alpha-helix (F). The Ca2+ ion is bound in the EF2–4 loops at specific sites (arrowhead); EF1 does not bind calcium. The mutations associated with a macular dystrophy (MD) phenotype are displayed with a hashtag. EF1–4, EF-hand domains; Myr, N-terminal myristoylation.
Clinical description of patients with GUCA1A mutations
Clinical evaluations of four generations in two families (MTP434 and MTP1560) were performed. They revealed an autosomal dominant pattern of inheritance (Figure 1A).
Family MTP434
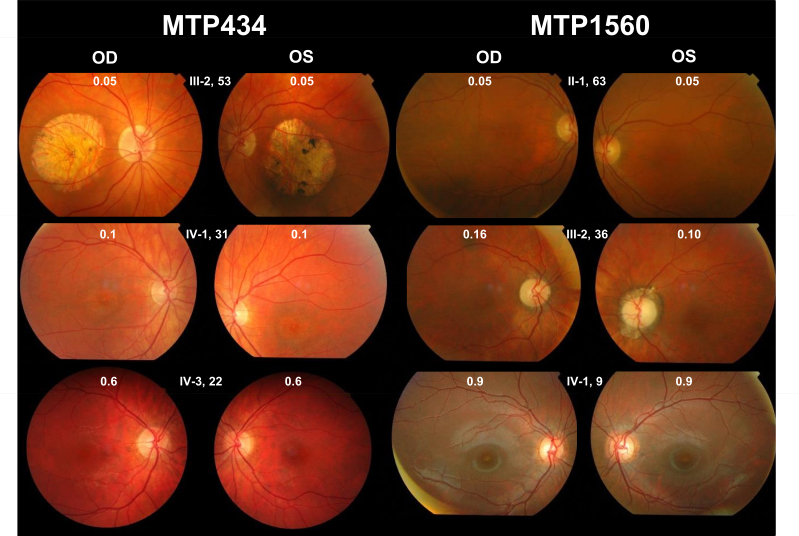
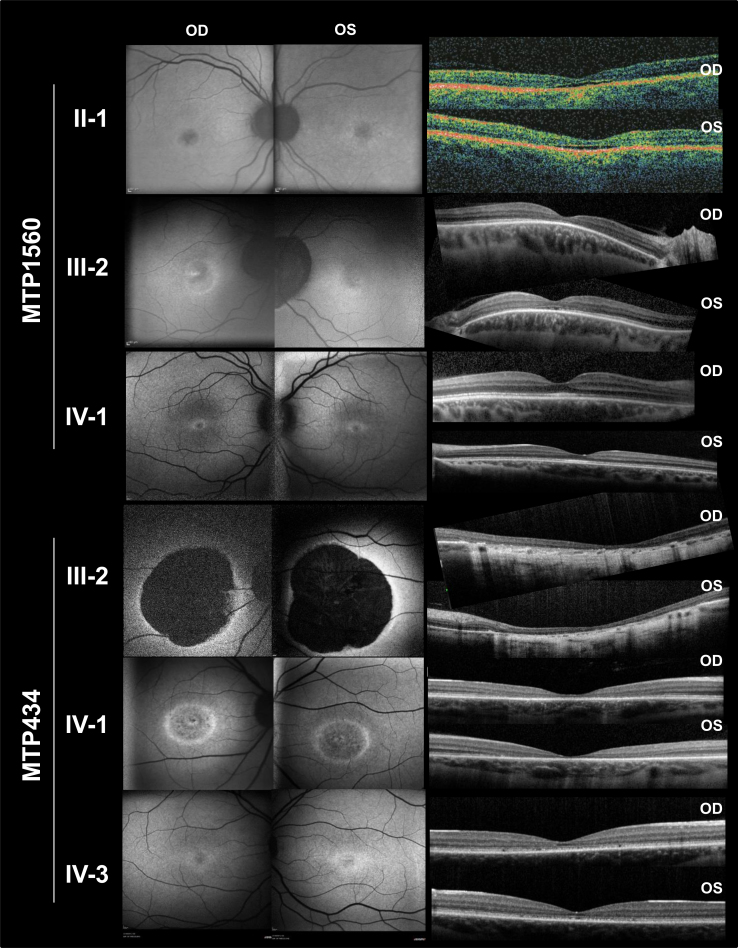
Three patients from the two last generations could be examined. They had all experienced the onset of visual acuity loss in adolescence, with moderate to severe reduction later in life; a perception of color vision impairment since childhood, with a variable axis of confusion (not shown); moderate night blindness in adulthood; and photophobia (Table 2). On visual-field examination, all had a central scotoma with preservation of the periphery (not shown). Fundus examination showed the absence of foveal reflex in the youngest patient (IV:3), perifoveal discoloration in his older sister (IV:1), and a large, round-shaped macular atrophy with sharp limits and few pigment deposits in the lesion in their mother (III:2; Figure 3). Fundus autofluorescence revealed a mild perifoveal hyperautofluorescence in IV:3, whereas IV:1 had a small, round hypoautofluorescent lesion, and III:2 exhibited no autofluorescence in the macular lesion (Figure 4). Importantly, in all patients, the retinal autofluorescence was normal outside the macular lesions. OCT examination confirmed complete (III:2) or near-complete (IV:1) foveal atrophy in two individuals, whereas in IV:3, a thinned ellipsoid zone was still present (Figure 4). Full-field ERG performed on the mother (III:2) and her daughter (IV:1) did not show significant alteration of the scotopic responses (Figure 5A). The photopic responses were not delayed. Their amplitudes were slightly decreased in III:2 and reduced to half the normal value in IV:1. Multifocal ERG could only be performed in two of the children. In IV:1, only the foveal responses had a decreased amplitude, whereas the responses outside the fovea were normal. In IV:3, most responses had a disorganized pattern except those of the outer ring, which showed normal amplitudes (Figure 5B). In summary, family MTP434 exhibited an MD phenotype.
Table 2. Clinical features of patients with GUCA1A mutations.
| Family patient | Age, sex | Symptoms | VA [refraction] OD VA [refraction] OS |
|---|---|---|---|
| MTP 434 III-2 |
53, F |
Photophobia, moderate night blindness onset at 25 |
0.05 [+2.25(−1,00)135°]
0.05 [+3.00(−1,00)65°] |
| MTP 434 IV-1 |
31, F |
Photophobia, moderate night blindness onset at 25 |
0.1 [+0.50(−0,50)160°]
0.1 [+0.25] |
| MTP 434 IV-3 |
22, M |
No night blindness, photophobia |
0.6 [-0.50]
0.6 [-0.50] |
| MTP 1560 II-1 |
63, M |
No night blindness, photophobia stopped driving at 45 |
0.05 [+4.75]
0.05 [+4.00(−0.50)135°] |
| MTP 1560 III-2 |
36, F |
No night blindness, photophobia |
0.16 [-2.50(−1,75)120°]
0.10 [-2.50(−1,25)165°] |
| MTP 1560 IV-3 | 9, M | No night blindness, photophobia | 0.9 [+2.50(−2,25)115°] 0.9 [+2.75(−3,00)70°] |
VA, visual acuity; M, male; F, female; OD, oculus dexter (right eye); OS, oculus sinister (left eye).
Figure 3.
Fundus imaging of patients with autosomal dominant cone dystrophy (adCOD) or macular dystrophy (adMD) and with GUCA1A mutations. On top of each picture, the family number, patient number in the family, age, and visual acuity in decimal values are indicated. OD, oculus dexter (right eye); OS, oculus sinister (left eye). Family MTP434, absence of foveal reflex in IV:3; perifoveal discoloration in IV:1; and large, round-shaped macular atrophy with sharp limits and few pigment deposits in the lesion in III:2. Family MTP1560, slight decreased foveal reflex in IV:1 and absence of foveal reflex in III:2 and in II:1.
Figure 4.
Fundus autofluorescence (FAF) photographs and spectral-domain optical coherence tomography (OCT) scans of patients with GUCA1A mutations. Family MTP434 FAF, mild perifoveal hyperautofluorescence in IV:3; small, round, hypoautofluorescent lesion in IV:1; and no autofluorescence in the macular lesion in III:2. Family MTP434 OCT, complete (in IV:1) or near-complete (in III:2) foveal atrophy; a thinned ellipsoid zone remained present in IV:3. Family MTP1560 FAF, moderate perifoveal autofluorescence in IV:1, fovea moderately hypoautofluorescent in III:2 and II:1, and normal autofluorescence in the peripheral retina for the three patients. Family MTP1560 OCT, thickened ellipsoid zone in the fovea in IV:1, absent ellipsoid zone with thinning of the outer nuclear layer in the fovea in III:2, and partial foveal atrophy with a hyporeflective zone in both eyes in II:1. OD, oculus dexter (right eye); OS, oculus sinister (left eye).
Figure 5.
Full-field electroretinography (ffERG) and multifocal ERG (mfERG) of patients with GUCA1A mutations. A: Family MTP434 ffERG, no significant alteration of the scotopic responses in III:2 and IV:1, slight decrease of the photopic responses in III:2, and reduction to half the value in IV:1. Family MTP1560 ffERG, normal dim blue scotopic responses and moderate decrease of the bright white scotopic responses in all patients. Dramatic decrease of the photopic responses in all patients, although slightly better in IV:1, and 30-Hz flickers delayed. B: Family MTP434 mfERG, foveal decreased amplitude in IV:1 and normal amplitudes of the outer ring only in IV:3. Family MTP1560 mfERG, highly abnormal responses in IV:1 and in III:2.
Family MTP1560
Patients from the three last generations could be examined. They all complained of photophobia and color vision difficulties that had been present since childhood, but none reported night blindness. Visual acuity was variously decreased (Table 2). None of the patients had macular atrophy; the son (IV:1) had a slightly decreased foveal reflex that was absent in his mother (III:2) and his grandfather (II:1; Figure 3). Fundus autofluorescence showed small foveal lesions in the three generations and normal autofluorescence in the peripheral retina; moderate perifoveal hyperautofluorescence was observed in IV:1, whereas the fovea was moderately hypoautofluorescent in III:2 and II:1 (Figure 4). OCT examination of the son (IV:1) revealed a thickened ellipsoid zone in the fovea (Figure 4). His mother (III:2) had an absent ellipsoid zone, with thinning of the outer nuclear layer in the fovea, and a virtually normal outer retina outside the fovea in the temporal area of the macula. The grandfather (II:1) had partial foveal atrophy, with a hyporeflective zone in both eyes. Full-field dim blue scotopic ERG responses were normal in the three family members (Figure 5A), but the scotopic responses for the brightest stimulation were moderately decreased. In the three family members, the photopic responses were dramatically decreased, although they were slightly better in IV:1, and the 30-Hz flickers were delayed. Multifocal ERG, performed for IV:1 and his mother (III:2), showed highly abnormal responses (Figure 5B). In III:2, all responses were disorganized. In IV:1, most responses showed decreased amplitudes. In summary, family MTP1560 exhibited a COD phenotype.
Discussion
To date, 16 different mutations in GUCA1A that lead to adCOD, adCORD or adMD have been listed on the Human Genome Mutation Database (HGMD), except the unclear p.Thr114Ile substitution associated with atypical RP (Table 1). In this study, we report two novel mutations in GUCA1A and clinical data associated with the two families in which the mutations were identified, where one had adCOD and the other had adMD. The two novel mutations affect evolutionarily conserved amino acids (Figure 1C) and are located in a highly-conserved region, namely the EF-hand domains of the GCAP1 protein, which contain most disease-causing mutations (Table 1, Figure 2). The helix–loop–helix structure, called the EF hand, was first identified in parvalbumin, where the two helices E and F, joined by a calcium-binding loop, resemble a hand [26]. This EF-hand motif is found in many calcium-binding proteins. The GCAP1 loop region of the EF2–4 hands contains 12 residues, and several of these residues are involved in the binding of the Ca2+ (Figure 2, arrowheads). The EF1 hand is not capable of binding Ca2+.
Among the 16 causative mutations in GUCA1A referenced in the literature, six affected one of the amino acids directly involved in the binding of the Ca2+ in the EF loop domains (Figure 2; p.Asp100Glu, p.Asp100Gly, p.Asp102His, p.Asn104Lys, p.Glu155Gly, and p.Glu155Ala). For the mutation p.Asp100Glu in the EF3-hand motif, it was shown that although the exchange of Asp for Glu at position 100 added only a CH2 group to the side chain, it caused an impaired apparent Ca2+ affinity, which was 100-fold lower than that of the wild type [27,28]. The novel mutation p.Asp148Glu, which affects one of the Ca2+-binding EF4 amino acids with a concomitant exchange of Asp for Glu at position 148, probably lowers the affinity of the EF4 to Ca2+. The mutation p.Val101del is located at a position that is not directly involved in complexing Ca2+, although it appears between two Ca2+-binding aspartic acid residues at positions 100 and 102 of the EF3 hand (Figure 2). The in-frame deletion of one valine at position 101 may affect the loop conformation and the Ca2+ binding on the adjacent aspartic acid residues.
All the GUCA1A mutations listed in Table 1 (the two novel mutations of the present study and the 16 referenced mutations) are absent from the public genetic databases except the following two: p.Pro50Leu, found at a high frequency (125/121,374 alleles in ExAC and 13/13,006 alleles in EVS), and p.Thr114Ile (5/121,228 alleles in ExAC). This means that 125 carriers of p.Pro50Leu and 5 carriers of p.Thr114Ile were identified in an ExAC cohort comprising more than 60,000 individuals from the general population. The prevalence of the condition due to p.Pro50Leu and p.Thr114Ile would be 1 in 485 and 1 in 12,122 individuals respectively, which is more than the estimated prevalence for COD and CORD including all inheritance patterns (1 in 40,000 individuals) [1]. The variations p.Thr114Ile and p.Pro50Leu affect amino acids that are not evolutionarily conserved (Figure 1C and data not shown, respectively). Furthermore, the family described with p.Pro50Leu presented intrafamilial variability in expression [29]. The mutation p.Thr114Ile was identified in a simplex patient with an atypical form of retinal degeneration, but no cosegregation analysis was possible to validate the mutation [30]. Unfortunately, the mutated protein was not biochemically characterized in the study. Moreover, p.Pro50Leu and p.Thr114Ile are predicted to be benign by PolyPhen2 and SIFT, and the latter is also predicted benign by PROVEAN and MutationTaster (Table 1). In conclusion, these two variants are unlikely causative mutations of adCOD, adCORD, or adMD.
The present study found the prevalence of GUCA1A to be 16.7% in our small cohort of 12 families. Previous prevalence studies conducted in patients with adCOD and adCORD revealed that among the 10 disease-causing genes, each gene prevalence is low, but together, they explain approximately 20% of cases [31]. The estimated frequency of these genes is 7.7% for GUCY2D, 4.3% for CRX, 3.0% for GUCA1A, 1.8% for PRPH2, 1.4% for PROM1, and 1.2% for PITPNM3. The last four genes (AIPL1, RIMS, SEMA4A, and UNC119) are mutated in less than 1% of patients [31]. In the literature, the prevalence of GUCA1A mutations varies widely from 0.76% to 16.7% for cases of adCOD and adCORD in cohorts of different origins. In a cohort of 131 patients with cone degeneration, cone–rod degeneration, or cone dysfunction, only one patient had a mutation in GUCA1A (0.76%) [30]. In two characterized cohorts of 22 and 24 patients with adCOD and adCORD, respectively, 3/22 (13.6%) [27] and 4/24 (16.7%) [32] patients had a causative mutation in GUCA1A; these frequencies are similar to those obtained in our study.
For family MTP1560, the phenotype of the patients with p.Asp148Glu mutation was that of COD on full-field ERG, and none of the patients had macular atrophy. Conversely, the patients in family MTP434 with the p.Val101del mutation had MD. COD and CORD were first associated with GUCA1A mutations [30,33-35]. Next, six referenced GUCA1A mutations were associated with an MD phenotype (Table 1), namely p.Leu84Phe [36], p.Glu89Lys [27], Tyr99Cys [37], p.Asp100Gly [38], p.Ile107Thr [36], and p.Leu176Phe [39]. The first report of a macular dysfunction caused by the GUCA1A mutation was published by Michaelides and collaborators [37]. They described six affected individuals from a family with the p.Tyr99Cys mutation displaying isolated MD, COD, and CORD phenotypes. Then, one family with p.Glu89Lys mutation carried by two patients, one with an unknown clinical status and one with COD or MD, was reported [27]. After that, two Spanish families, including a three-generation family with five affected patients carrying the p.Leu84Phe mutation and one index patient with the p.Ile107Thr mutation, were described with intrafamilial heterogeneity that included isolated macular dysfunction, COD, and CORD [36]. Thereafter, a family of three affected individuals with the p.Asp100Gly mutation was described with a heterogeneous presentation of phenotypes and macular atrophy, cone, and cone–rod dysfunction on ERG [38]. Finally, a missense variation, p.Leu176Phe, was identified via whole-exome sequencing in a patient with MD [39]. No information about the clinical status of the patient or about the cosegregation on additional samples were presented. Except for this latter mutation, all the five referenced mutations and the novel p.Val101del associated with MD were localized from the EF2 helix F to the EF3 loop (Figure 2, hashtag). Contrary to what is presented in the literature for the MD phenotype associated with GUCA1A mutations, there was no intrafamilial phenotypic heterogeneity in family MTP434 with the p.Val101del mutation. The three patients examined had MD and no signs of COD or CORD.
Biochemical analysis of GCAP1 function revealed that for most mutations identified in GUCA1A, the Ca2+-sensor GCAP1 exhibits an impaired Ca2+ sensitivity and a dysfunction of its guanylate cyclase–activating properties, as summarized in two reviews [10,40]. Based on these biochemical analysis, we hypothesized that the two novel GUCA1A mutations described in this study, p.Val101del and p.Asp148Glu, result in defects in the GCAP1 EF hands and affect calcium homeostasis. This resulted in MD for p.Val101del and COD for p.Asp148Glu.
Acknowledgments
No financial disclosures. The authors acknowledge funding support from the SOS Rétinite Foundation (Montpellier, France) for a fellowship to GM.
Appendix 1.
To access the data, click or select the words “Appendix 1.” For each family, the number of affected generations and affected patients are indicated. COD, cone dystrophy; CORD, cone-rod dystrophy; MD, macular dystrophy; ad, autosomal dominant.
Appendix 2. Sequence of the primers (5'-3'), fusion temperatures and concentration of MgCl2 used to amplify GUCA1A (NM_000409.3)
To access the data, click or select the words “Appendix 2.” Amplification condition was performed in a 25 µl reaction volume and consisting of 16.8 µl nuclease free water, 2.5 µl of TAQ polymerase buffer, 1.5 or 2 µl MgCl2, 0,5 µl forward primer, 0,5 µl reverse primer, 0.2 µl TAQ polymerase, 2 µl template. Thermal cycle programmed for 90 s at 95 °C as initial denaturation, followed by 35 cycles of 30 s at 95 °C for denaturation, 30 s at 60 °C or 62 °C as annealing, 60 s at 72 °C for extension, and final extension at 72 °C for 10 min. PCR products were examined by electrophoresis at 100 V for 30 min in a 1.5% (w/v) agarose gel in 1X TAE buffer. The marker used DNA ladder 100 bp. Electrophoresis gel was soaked in ethidium bromide for 30 min then visualized in UV light.
References
- 1.Hamel CP. Cone rod dystrophies. Orphanet J Rare Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simunovic MP, Moore AT. The cone dystrophies. Eye (Lond) 1998;12(Pt 3b):553–65. doi: 10.1038/eye.1998.145. [DOI] [PubMed] [Google Scholar]
- 3.Thiadens AAHJ, Phan TML, Zekveld-Vroon RC, Leroy BP, van den Born LI, Hoyng CB, Klaver CCW, Writing Committee for the Cone Disorders Study Group Consortium Roosing S, Pott J-WR, van Schooneveld MJ, van Moll-Ramirez N, van Genderen MM, Boon CJF, den Hollander AI, Bergen AAB, De Baere E, Cremers FPM, Lotery AJ. Clinical course, genetic etiology, and visual outcome in cone and cone-rod dystrophy. Ophthalmology. 2012;119:819–26. doi: 10.1016/j.ophtha.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Olshevskaya EV, Ermilov AN, Dizhoor AM. Factors that affect regulation of cGMP synthesis in vertebrate photoreceptors and their genetic link to human retinal degeneration. Mol Cell Biochem. 2002;230:139–47. [PubMed] [Google Scholar]
- 5.Palczewski K, Polans AS, Baehr W, Ames JB. Ca(2+)-binding proteins in the retina: structure, function, and the etiology of human visual diseases. BioEssays. 2000;22:337–50. doi: 10.1002/(SICI)1521-1878(200004)22:4<337::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 6.Arshavsky VY, Burns ME. Photoreceptor signaling: supporting vision across a wide range of light intensities. J Biol Chem. 2012;287:1620–6. doi: 10.1074/jbc.R111.305243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chabre M, Deterre P. Molecular mechanism of visual transduction. Eur J Biochem. 1989;179:255–66. doi: 10.1111/j.1432-1033.1989.tb14549.x. [DOI] [PubMed] [Google Scholar]
- 8.Yarfitz S, Hurley JB. Transduction mechanisms of vertebrate and invertebrate photoreceptors. J Biol Chem. 1994;269:14329–32. [PubMed] [Google Scholar]
- 9.Frins S, Bönigk W, Müller F, Kellner R, Koch KW. Functional characterization of a guanylyl cyclase-activating protein from vertebrate rods. Cloning, heterologous expression, and localization. J Biol Chem. 1996;271:8022–7. doi: 10.1074/jbc.271.14.8022. [DOI] [PubMed] [Google Scholar]
- 10.Koch K-W, Dell’orco D. A calcium-relay mechanism in vertebrate phototransduction. ACS Chem Neurosci. 2013;4:909–17. doi: 10.1021/cn400027z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palczewski K, Subbaraya I, Gorczyca WA, Helekar BS, Ruiz CC, Ohguro H, Huang J, Zhao X, Crabb JW, Johnson RS. Molecular cloning and characterization of retinal photoreceptor guanylyl cyclase-activating protein. Neuron. 1994;13:395–404. doi: 10.1016/0896-6273(94)90355-7. [DOI] [PubMed] [Google Scholar]
- 12.Kachi S, Nishizawa Y, Olshevskaya E, Yamazaki A, Miyake Y, Wakabayashi T, Dizhoor A, Usukura J. Detailed localization of photoreceptor guanylate cyclase activating protein-1 and −2 in mammalian retinas using light and electron microscopy. Exp Eye Res. 1999;68:465–73. doi: 10.1006/exer.1998.0629. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Seno K, Nishizawa Y, Hayashi F, Yamazaki A, Matsumoto H, Wakabayashi T, Usukura J. Ultrastructural localization of retinal guanylate cyclase in human and monkey retinas. Exp Eye Res. 1994;59:761–8. doi: 10.1006/exer.1994.1162. [DOI] [PubMed] [Google Scholar]
- 14.Yang RB, Foster DC, Garbers DL, Fülle HJ. Two membrane forms of guanylyl cyclase found in the eye. Proc Natl Acad Sci USA. 1995;92:602–6. doi: 10.1073/pnas.92.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory-Evans K, Kelsell RE, Gregory-Evans CY, Downes SM, Fitzke FW, Holder GE, Simunovic M, Mollon JD, Taylor R, Hunt DM, Bird AC, Moore AT. Autosomal dominant cone-rod retinal dystrophy (CORD6) from heterozygous mutation of GUCY2D, which encodes retinal guanylate cyclase. Ophthalmology. 2000;107:55–61. doi: 10.1016/s0161-6420(99)00038-x. [DOI] [PubMed] [Google Scholar]
- 16.Kelsell RE, Gregory-Evans K, Payne AM, Perrault I, Kaplan J, Yang RB, Garbers DL, Bird AC, Moore AT, Hunt DM. Mutations in the retinal guanylate cyclase (RETGC-1) gene in dominant cone-rod dystrophy. Hum Mol Genet. 1998;7:1179–84. doi: 10.1093/hmg/7.7.1179. [DOI] [PubMed] [Google Scholar]
- 17.Kitiratschky VBD, Wilke R, Renner AB, Kellner U, Vadalà M, Birch DG, Wissinger B, Zrenner E, Kohl S. Mutation analysis identifies GUCY2D as the major gene responsible for autosomal dominant progressive cone degeneration. Invest Ophthalmol Vis Sci. 2008;49:5015–23. doi: 10.1167/iovs.08-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payne AM, Morris AG, Downes SM, Johnson S, Bird AC, Moore AT, Bhattacharya SS, Hunt DM. Clustering and frequency of mutations in the retinal guanylate cyclase (GUCY2D) gene in patients with dominant cone-rod dystrophies. J Med Genet. 2001;38:611–4. doi: 10.1136/jmg.38.9.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dizhoor AM, Olshevskaya EV, Henzel WJ, Wong SC, Stults JT, Ankoudinova I, Hurley JB. Cloning, sequencing, and expression of a 24-kDa Ca(2+)-binding protein activating photoreceptor guanylyl cyclase. J Biol Chem. 1995;270:25200–6. doi: 10.1074/jbc.270.42.25200. [DOI] [PubMed] [Google Scholar]
- 20.Gorczyca WA, Polans AS, Surgucheva IG, Subbaraya I, Baehr W, Palczewski K. Guanylyl cyclase activating protein. A calcium-sensitive regulator of phototransduction. J Biol Chem. 1995;270:22029–36. doi: 10.1074/jbc.270.37.22029. [DOI] [PubMed] [Google Scholar]
- 21.Imanishi Y, Li N, Sokal I, Sowa ME, Lichtarge O, Wensel TG, Saperstein DA, Baehr W, Palczewski K. Characterization of retinal guanylate cyclase-activating protein 3 (GCAP3) from zebrafish to man. Eur J Neurosci. 2002;15:63–78. doi: 10.1046/j.0953-816x.2001.01835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuenca N, Lopez S, Howes K, Kolb H. The localization of guanylyl cyclase-activating proteins in the mammalian retina. Invest Ophthalmol Vis Sci. 1998;39:1243–50. [PubMed] [Google Scholar]
- 23.Otto-Bruc A, Fariss RN, Haeseleer F, Huang J, Buczyłko J, Surgucheva I, Baehr W, Milam AH, Palczewski K. Localization of guanylate cyclase-activating protein 2 in mammalian retinas. Proc Natl Acad Sci USA. 1997;94:4727–32. doi: 10.1073/pnas.94.9.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howes K, Bronson JD, Dang YL, Li N, Zhang K, Ruiz C, Helekar B, Lee M, Subbaraya I, Kolb H, Chen J, Baehr W. Gene array and expression of mouse retina guanylate cyclase activating proteins 1 and 2. Invest Ophthalmol Vis Sci. 1998;39:867–75. [PubMed] [Google Scholar]
- 25.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kretsinger RH, Nockolds CE. Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem. 1973;248:3313–26. [PubMed] [Google Scholar]
- 27.Kitiratschky VBD, Behnen P, Kellner U, Heckenlively JR, Zrenner E, Jägle H, Kohl S, Wissinger B, Koch K-W. Mutations in the GUCA1A gene involved in hereditary cone dystrophies impair calcium-mediated regulation of guanylate cyclase. Hum Mutat. 2009;30:E782–96. doi: 10.1002/humu.21055. [DOI] [PubMed] [Google Scholar]
- 28.Dell’Orco D, Behnen P, Linse S, Koch K-W. Calcium binding, structural stability and guanylate cyclase activation in GCAP1 variants associated with human cone dystrophy. Cell Mol Life Sci. 2010;67:973–84. doi: 10.1007/s00018-009-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Downes SM, Holder GE, Fitzke FW, Payne AM, Warren MJ, Bhattacharya SS, Bird AC. Autosomal dominant cone and cone-rod dystrophy with mutations in the guanylate cyclase activator 1A gene-encoding guanylate cyclase activating protein-1. Arch Ophthalmol. 2001;119:96–105. [PubMed] [Google Scholar]
- 30.Nishiguchi KM, Sokal I, Yang L, Roychowdhury N, Palczewski K, Berson EL, Dryja TP, Baehr W. A Novel Mutation (I143NT) in Guanylate Cyclase-Activating Protein 1 (GCAP1) Associated with Autosomal Dominant Cone Degeneration. IOVS. 2004;45:3863–70. doi: 10.1167/iovs.04-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roosing S, Thiadens AAHJ, Hoyng CB, Klaver CCW, den Hollander AI, Cremers FPM. Causes and consequences of inherited cone disorders. Prog Retin Eye Res. 2014;42:1–26. doi: 10.1016/j.preteyeres.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Kitiratschky VBD, Glöckner CJ, Kohl S. Mutation screening of the GUCA1B gene in patients with autosomal dominant cone and cone rod dystrophy. Ophthalmic Genet. 2011;32:151–5. doi: 10.3109/13816810.2011.559650. [DOI] [PubMed] [Google Scholar]
- 33.Payne AM, Downes SM, Bessant DA, Taylor R, Holder GE, Warren MJ, Bird AC, Bhattacharya SS. A mutation in guanylate cyclase activator 1A (GUCA1A) in an autosomal dominant cone dystrophy pedigree mapping to a new locus on chromosome 6p21.1. Hum Mol Genet. 1998;7:273–7. doi: 10.1093/hmg/7.2.273. [DOI] [PubMed] [Google Scholar]
- 34.Sokal I, Dupps WJ, Grassi MA, Brown J, Jr, Affatigato LM, Roychowdhury N, Yang L, Filipek S, Palczewski K, Stone EM, Baehr W. A novel GCAP1 missense mutation (L151F) in a large family with autosomal dominant cone-rod dystrophy (adCORD). Invest Ophthalmol Vis Sci. 2005;46:1124–32. doi: 10.1167/iovs.04-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkie SE, Li Y, Deery EC, Newbold RJ, Garibaldi D, Bateman JB, Zhang H, Lin W, Zack DJ, Bhattacharya SS, Warren MJ, Hunt DM, Zhang K. Identification and functional consequences of a new mutation (E155G) in the gene for GCAP1 that causes autosomal dominant cone dystrophy. Am J Hum Genet. 2001;69:471–80. doi: 10.1086/323265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamenarova K, Corton M, García-Sandoval B, Fernández-San Jose P, Panchev V, Avila-Fernández A, López-Molina MI, Chakarova C, Ayuso C, Bhattacharya SS. Novel GUCA1A mutations suggesting possible mechanisms of pathogenesis in cone, cone-rod, and macular dystrophy patients. BioMed Res Int. 2013;2013:517570. doi: 10.1155/2013/517570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michaelides M, Wilkie SE, Jenkins S, Holder GE, Hunt DM, Moore AT, Webster AR. Mutation in the gene GUCA1A, encoding guanylate cyclase-activating protein 1, causes cone, cone-rod, and macular dystrophy. Ophthalmology. 2005;112:1442–7. doi: 10.1016/j.ophtha.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 38.Nong E, Lee W, Merriam JE, Allikmets R, Tsang SH. Disease progression in autosomal dominant cone-rod dystrophy caused by a novel mutation (D100G) in the GUCA1A gene. Doc Ophthalmol. 2014;128:59–67. doi: 10.1007/s10633-013-9420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisschuh N, Mayer AK, Strom TM, Kohl S, Glöckle N, Schubach M, Andreasson S, Bernd A, Birch DG, Hamel CP, Heckenlively JR, Jacobson SG, Kamme C, Kellner U, Kunstmann E, Maffei P, Reiff CM, Rohrschneider K, Rosenberg T, Rudolph G, Vámos R, Varsányi B, Weleber RG, Wissinger B. Mutation Detection in Patients with Retinal Dystrophies Using Targeted Next Generation Sequencing. PLoS One. 2016;11:e0145951. doi: 10.1371/journal.pone.0145951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behnen P, Dell’Orco D, Koch K-W. Involvement of the calcium sensor GCAP1 in hereditary cone dystrophies. Biol Chem. 2010;391:631–7. doi: 10.1515/BC.2010.063. [DOI] [PubMed] [Google Scholar]
- 41.Huang L, Xiao X, Li S, Jia X, Wang P, Sun W, Xu Y, Xin W, Guo X, Zhang Q. Molecular genetics of cone-rod dystrophy in Chinese patients: New data from 61 probands and mutation overview of 163 probands. Exp Eye Res. 2016;146:252–8. doi: 10.1016/j.exer.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 42.Jiang L, Wheaton D, Bereta G, Zhang K, Palczewski K, Birch DG, Baehr W. A novel GCAP1(N104K) mutation in EF-hand 3 (EF3) linked to autosomal dominant cone dystrophy. Vision Res. 2008;48:2425–32. doi: 10.1016/j.visres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang L, Li S, Xiao X, Jia X, Sun W, Gao Y, Li L, Wang P, Guo X, Zhang Q. Novel GUCA1A mutation identified in a Chinese family with cone-rod dystrophy. Neurosci Lett. 2013;541:179–83. doi: 10.1016/j.neulet.2013.02.013. [DOI] [PubMed] [Google Scholar]