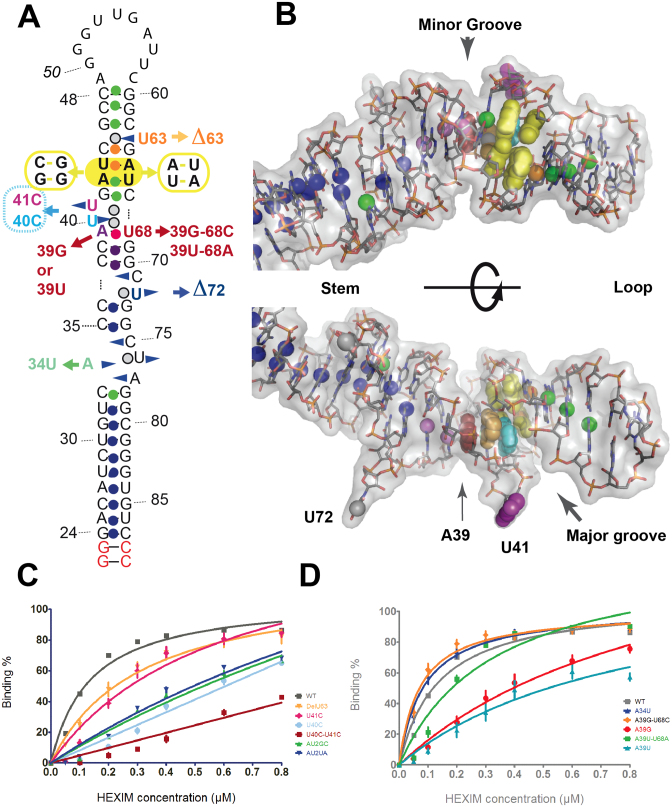
Figure 6.
HEXIM-binding study. (A) 2D sequence showing the mutations studied. The two terminal base-pairs (red) were added in the present analysis to ensure efficient production of the folded hairpin. The spheres represent the imino protons, colored according to their sensitivity to titration with the ARM-peptide (Lebars et al. 2010): no change (dark blue), binding (green), increase of solvent accessibility (orange), decrease of solvent accessibility (purple, red for A39-U68); not tested in the NMR experiment (grey). (B) Two views of hairpin HP1 (conformation OUT) with the HEXIM-binding site colored as in A. (C and D) Binding to HEXIM of the HP1 mutants measured with electrophoretic mobility shift assays. The experimental data (average of at least three experiments) were fitted with a model corresponding to one binding site. (C) Binding observed for mutations at the 7SK motif and (D) below the 7SK-motif.