Abstract
Site-directed A-to-I RNA editing is a technology for re-programming genetic information at the RNA-level. We describe here the first design of genetically encodable guideRNAs that enable the re-addressing of human ADAR2 toward specific sites in user-defined mRNA targets. Up to 65% editing yield has been achieved in cell culture for the recoding of a premature Stop codon (UAG) into tryptophan (UIG). In the targeted gene, editing was very specific. We applied the technology to recode a recessive loss-of-function mutation in PINK1 (W437X) in HeLa cells and showed functional rescue of PINK1/Parkin-mediated mitophagy, which is linked to the etiology of Parkinson's disease. In contrast to other editing strategies, this approach requires no artificial protein. Our novel guideRNAs may allow for the development of a platform technology that requires only the administration or expression of a guideRNA to recode genetic information, with high potential for application in biology and medicine.
INTRODUCTION
RNA editing alters genetic information at the RNA-level by insertion, deletion or modification of nucleotides (1). The catalytic deamination of adenosine (A) gives inosine (I) that is biochemically read as guanosine. In consequence, A-to-I RNA editing alters the function of RNAs in various ways. Amino acids are substituted, miRNA recognition (2,3) and splicing (4) are altered. In the human transcriptome, the classic example for amino acid substitution is the editing of the glutamate receptor GluR2 transcript at two sites, the R/G and the Q/R site, with the latter one being essential for nervous system function (5,6). A-to-I editing is carried out by two ADARs (adenosine deaminases acting on RNA), ADAR1 and ADAR2 in different isoforms (7). An alteration of RNA editing is linked to various neurological diseases including behavioral disorders, epilepsy and the Prader–Willi syndrome (8–11). Knock-out of ADAR2 in mice leads to an early dead of the newborn due to seizures. Interestingly, this phenotype can be rescued by genomic insertion of an R at the Q/R site in GluR2 (12). Mutations in ADAR1 are linked to the Aicardi–Goutieres syndrome, (13) an autoimmune disease and others, including dyschromatosis (14). Knock-out of ADAR1 function in mice results in early embryonic fatality, (15,16) which can be rescued by a simultaneous knock-out of dsRNA sensing via MDA5 (17,18). Both, hyper (19)- and hypoediting (20) have been associated with cancer (21–23). Whereas ADAR1 is ubiquitously expressed in various tissues, ADAR2 is mainly expressed in neurons (24). Both enzymes are promiscuously recruited to hundreds of thousands of double-stranded RNA structures by their N-terminal dsRNA binding domains (dsRBD) (25,26). Thus, editing is relatively unspecific, happens massively in Alu repeats, and on dsRNA structures of >30 bp (27). However, some RNA substrates, containing bulges and loops, are edited in a highly specific manner (7). In particular, the precise and efficient editing at the R/G site of the GluR2 transcript results from a defined positioning of ADAR2 by the interaction of its two dsRBDs with the exon/intron border of the transcript (28).
Re-directing RNA editing to user-defined targets allows altering genetic information in a highly rational way (29). Various applications in basic biology and medicine are conceivable. Even though limited to A-to-I substitution, the scope is large. It includes the targeting of most polar amino acids (Gln, Arg, His, Tyr, Ser, Thr and others), which play essential roles in enzyme catalysis, signaling and posttranslational modification. Furthermore, stop, start, splicing signals and miRNA recognition sites can be manipulated. Thus, site-directing RNA editing at specific sites on user-defined targets has an immense potential for the manipulation of protein function, RNA processing, and could be used to attenuate disease phenotypes (29). Such a strategy would complement current genome editing approaches (30) in several aspects. The transient and thus reversible nature of RNA manipulation could be beneficial with respect to ethical issues and safety aspects. Both, therapeutic and potential adverse effects are likely to be tunable and reversible. Furthermore, manipulations are conceivable that are inaccessible at the genome level. This includes amino acid or transcript level changes that would kill a cell if they are permanently enforced. Potentially lethal interventions on kinases, apoptosis factors, coagulation factors, (31) transcription or translation factors could be realized on the RNA-level suddenly, transiently or partially to obtain a therapeutic effect. Manipulation at the RNA-level might also be much more efficient compared to HDR-dependent gene correction, which remained persistently inefficient in vivo, (30) in particular in postmitotic tissues like the brain. For many genetic diseases caused by recessive loss-of-function mutations, a drug that can restore a small fraction (like 5%) of functional gene product in a large fraction of a tissue is superior to a drug that can restore full gene function (100%) but only in a small fraction of the tissue. A typical example is cystic fibrosis (32).
Site-directed RNA editing is not a new concept. The first trial goes back to the pioneering work of Tod Woolf and colleagues who could demonstrate already in 1995 to elicit RNA editing in a reporter mRNA inside Xenopus eggs when the mRNA was hybridized with a 52 nt long, unstructured guideRNA prior to microinjection (33,29). However, the main issue of low efficiency and in particular off-site editing in the guideRNA/mRNA could not be solved at that time. In 2012 and 2013, we (34,35) and the lab of Joshua Rosenthal (36) have reanimated the concept of site-directed RNA editing by independent engineering of artificial editing enzymes that address the catalytic activity in a highly rational way with the help of external guideRNAs. Such strategies work inside mammalian cell culture and even in a simple organism (37) and allow the repair of disease-relevant genes, like the CFTR (36) mRNA. Chemical modification of the guideRNA was shown to improve specificity (35). Even though feasible and expandable for multiplexing approaches, both established strategies require the expression of an engineered deaminase. With respect to this limitation, we were wondering if it may become possible to harness the endogenous human ADARs for site-directed RNA editing, again with external guideRNAs. As ADARs are well expressed in neurons, (24) such a strategy could enable the attenuation of (neuron-related) disease phenotypes related to loss-of-function mutations simply by administration or ectopic expression of a small guideRNA.
Neurodegenerative diseases are a global challenge of tomorrow (38). Their enormous costs in healthcare threaten the welfare system. Parkinson΄s disease (PD) affects the central nervous system, destroys motion control, is often accompanied by neuropsychiatric disorders and characterized by a slow progression (39). The disease results from a loss of dopaminergic neurons in the substantia nigra and is typically accompanied by the formation of Lewy bodies. Several genes are linked to inherited forms of PD including numerous mutations in α-synuclein, Parkin, PINK1 or LRRK2. However, some forms of hereditary early-onset forms of PD are linked to single mutations in one gene, like W437X in PINK1 (40). Studying such mutations has proven valuable for the elucidation of the mechanism underlying specific forms of PD. Recent research shows that PINK1 and Parkin work together in a mitochondria quality control pathway, where damaged depolarized mitochondria are eliminated by the process of autophagy, termed mitophagy. The PINK1 kinase-function is required in an initial and essential step of mitophagy, which is the recruitment of cytosolic Parkin to the damaged mitochondria and the formation of perinuclear clusters (41). A single G-to-A nucleotide exchange in PINK1 has been described that changes Trp437 into a premature Stop codon and truncates PINK1's C-terminus by 145 amino acids including the functionally important kinase domain (40). This results in the impairment of the Parkin-dependent perinuclear clustering, clearance of damaged mitochondria and is linked to early-onset PD.
Here, we describe the rationale for the design of guideRNAs to harness human ADAR2 for site-directed RNA editing. We demonstrate the feasibility of the approach by the repair of a neuron-related disease-causing point mutation and show functional rescue of a mitophagy phenotype.
MATERIALS AND METHODS
Protein production
Wild-type human ADAR2 (with a C-terminal His6-tag) was produced from yeast (YVH10), purified by nickel and heparin affinity chromatography similar as described before (42). For details see Supplementary Material.
R/G-guideRNA synthesis
R/G-guideRNAs were produced by T7 in vitro transcription. The guideRNAs were cleaved during transcription from a cis-acting hammerhead ribozyme (TA TTCCACCTGA TGAGTTTTTA CGAAACGTTC CCGTGAGGGA ACGTC*GTGGAATA, the guideRNA starts after the asterisk that marks the cleavage site). The guideRNAs were purified by urea (7.5M) PAGE (8%, 1xTBE), isolated by the crush soak method and precipitated with ethanol.
In vitro editing
Editing assays were performed with purified mRNAs, guideRNAs and ADAR2-His6 protein. mRNA (0.5 or 25 nM), guideRNA (5 or 125 nM) and ADAR2 (180 or 350 nM) were incubated in reverse transcription buffer (75 mM KCl, 25 mM Tris-HCl, 2 mM DTT, pH 8.3) and the indicated amounts of magnesium (1.5 or 3 mM), and spermidine (0, 0.5 or 2 mM). Editing reactions were cycled three times between 37°C (30 min) and 30°C (30 min) and were stopped by addition of a sense oligomer that displaces the guideRNA from the mRNA. After reverse transcription and Taq-PCR, DNA was analyzed by Sanger sequencing. The editing yields were estimated by the relative areas of the guanosine versus adenosine traces.
Cellular editing
(a) under transient ADAR2 expression: ADAR2 (lacking the His-tag) was subcloned into the pcDNA3.1 vector as described in the Supplementary Material. The R/G guideRNAs were subcloned into the pSilencer2.1-U6hygro vector under control of U6 promoter and terminator. The W58X eGFP gene was cloned into the pcDNA3.1 vector as described before (29). 293T cells were cultivated with Dulbecco's modified Eagle's medium (DMEM), 10% fetal bovine serum (FBS), 1% P/S, 37°C, 5% CO2. Cells (1.75 × 105/well) were seeded into 24-well plates and were 24 h later transfected with the indicated amounts of plasmids using Lipofectamine 2000. Plasmid amounts have typically been 300 ng ADAR2, 300 ng W58X GFP, 1300–1600 ng guideRNA per well. For details see Supplementary Figures S8–S15. After the indicated time after transfection (typically 48 h) editing was evaluated by fluorescence microscopy (Zeiss AxioObserver.Z1) and RNA sequencing. For the latter, total RNA from the cells (Qiagen RNeasy Mini Kit) was DNaseI-digested, followed by reverse transcription, Taq-PCR amplification and Sanger sequencing. (b) under genomic ADAR2 expression: the 293 Flp-In T-REx system (Life Technologies) was used for stable integration of a single copy of ADAR2 at a genomic FRT-site in the cells. Briefly, 4 × 106 cells were seeded on a 10 cm dish. After one day, 1 μg of ADAR2 in a pcDNA5 vector under control of the tet-on CMV promoter, and 9 μg of pOG44 expressing the Flp recombinase were transfected with Lipofectamine 2000 (30 μl). One day later, the medium was changed for at least two weeks to selection medium (DMEM, 10% FBS, 100 μg/ml hygromycin B, 15 μg blasticidin S). Cells were kept in selection medium prior to the editing experiment which was then done in the absence of antibiotics. Editing: 3 × 105/well were seeded in poly-d-lysin-coated 24-well plates. Twenty four hours later, transfection was performed with GFP (300 ng) and R/G-guideRNA (1300 ng) using Lipofectamine 2000. GFP phenotype was analyzed by fluorescence microscopy. RNA was isolated and sequenced as described above 72 h post transfection. The sequences of all guideRNAs are given in Supplementary Table S1.
Mitophagy assay
HeLa cells (PINK1 wt or KO) were cultured under standard conditions (DMEM + 10% FBS, 37°C, 5% CO2). The mitophagy assay was performed in 24-well dishes. Each well contained a cover-slip coated with poly-d-lysine (Sigma Aldrich). The cells were seeded at 2.5 × 104/well. After 24 h, cells were transfected with the indicated plasmids using FuGene6 (Promega) following the manufacturer's instructions. If not indicated plasmid amounts/well were 300 ng for EGFP-Parkin, 300 ng for PINK W437amber and 300 ng editing vector. In control experiment Figure 4, d) 1300 ng of a guideRNA plasmid based on pSilencer lacking ADAR2 was co-transfected instead of the editing vector. In control experiment Figure 4, e), 200 ng of an editing vector lacking any guideRNAs but containing ADAR2 was co-transfected instead of the original editing vector. Treatment with 10 μM CCCP (in DMEM + 10% FBS) was either performed 46 h after transfection for 2 h or 24 h after transfection for 24 h. To visualize the mitochondria with a membrane potential sensitive dye, like MitoTracker Red CMXRos, a CCCP wash out was performed. For this, the depolarizing agent CCCP was washed out by changing the media twice every 15 min. Then the cells were incubated with 100 nM MitoTracker Red CMXROS (Invitrogen, M7512) in DMEM for 30 min at 37°C prior fixation or harvesting. Always 48 h after transfection, cells were washed with phosphate buffered saline (PBS) and then either fixated (4% paraformaldehyde/PBS) for immunocytochemical staining (A) or harvested for RNA isolation (B). (A) For immunocytochemical staining, the cells were washed 3 times with PBS and permeabilized with 1% Triton-X-100 in PBS for 5 min. After additional 3 washing steps, the cells were blocked in 10% FCS in PBS for 1 h at RT and incubated with following antibodies for 2 h at RT: mouse anti-ADAR2 (Santa Cruz, sc-73409) and rabbit anti-PINK1 (Novus Biological, BC-100-494). Subsequently, the cells were incubated with secondary antibodies: goat anti-rabbit or anti-mouse Alexa Fluor 568, 647 or 350. The nuclei were counterstained with Hoechst33342 in PBS. The cover-slips were mounted on glass-slides using Dako fluorescent mounting medium. The cells were analyzed using an Axio-Imager equipped with ApoTome (Zeiss). (B) RNA was isolated from the cells (Qiagen RNeasy Mini Kit). This was followed by DNaseI digest, reverse-transcription, amplification and Sanger sequencing. For further details see the Supplementary Material.
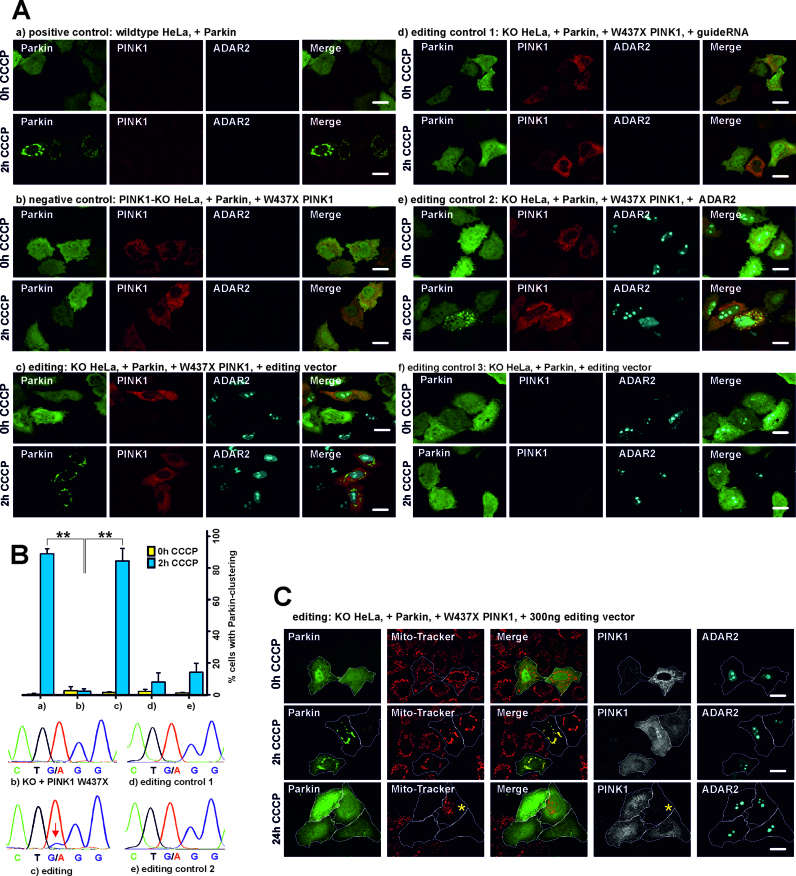
Figure 4.
Assaying PINK1 function. (A) Parkin-clustering during CCCP treatment. HeLa wild type or PINK1-KO cells were transfected with the respective plasmids and treated either with CCCP or DMSO (=CCCP 0 h) for 2 h or 24 h as indicated. Afterwards, PINK1 and ADAR2 were stained with antibodies, Parkin was tagged with EGFP for subsequent fluorescence microscopy. The scale bars indicate 20 μm. (B) Analysis of the editing experiments (a) to (e) shown in panel A. Top: quantification of cells with and without CCCP treatment for Parkin-clustering. The error bars give the standard deviation for N = 3 independent replications (always around 100 cells were counted). The asterisk(s) denote statistical significance when compared to sample (b) (2 h CCCP), with *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005. Bottom: Sanger sequencing of the PINK1 transcript after RT-PCR. Only sample (c) showed detectable editing as indicated by a red arrow. (a) to (f) in panel A and B correspond to the same experiments. (C) Restoration of the mitophagy phenotype by editing. After long-term CCCP treatment (24 h), triple positive cells (expressing Parkin, PINK1 and the editing vector) show clearance of the mitochondria that were visualized by Mito-Tracker (Red CMXRos) staining after CCCP washout (30 min). The Mito-Tracker and GFP channel have been merged to visualize the mitochondrial Parkin localization after 2 h CCCP treatment. For better visualization of mitophagy, the Parkin-positive cells have been encircled. The yellow asterisk (24 h CCCP treatment) marks an example of a cell that lacks PINK1 expression and hence did not perform mitophagy.
RESULTS AND DISCUSSION
Design of a trans-acting guideRNA
At the R/G-site of the natural GluR2 transcript, a cis-located intronic sequence folds back to the exon under formation of a bulged stem loop structure that recruits ADAR2 via its two dsRBDs (Figure 1A). A trans-acting guideRNA is conceivable that contains a part of the natural cis-acting R/G-motif in combination with an mRNA-binding platform to bind an arbitrary mRNA in order to recruit ADAR2 for site-directed editing. We designed a trans-acting guideRNA based on available structural information on the binding complex of dsRBD1 and 2 with the R/G-hairpin structure (28). We decided to cut the native R/G-site between the two guanosines five and six nucleotides downstream of the editing site (Figure 1A). This position appeared most suitable to harbor the protruding mRNA under minimal interference with dsRBD2 recognition and the deaminase domain. The latter assumption was confirmed by a recent crystal structure of the ADAR2 deaminase domain with an RNA model substrate (43).
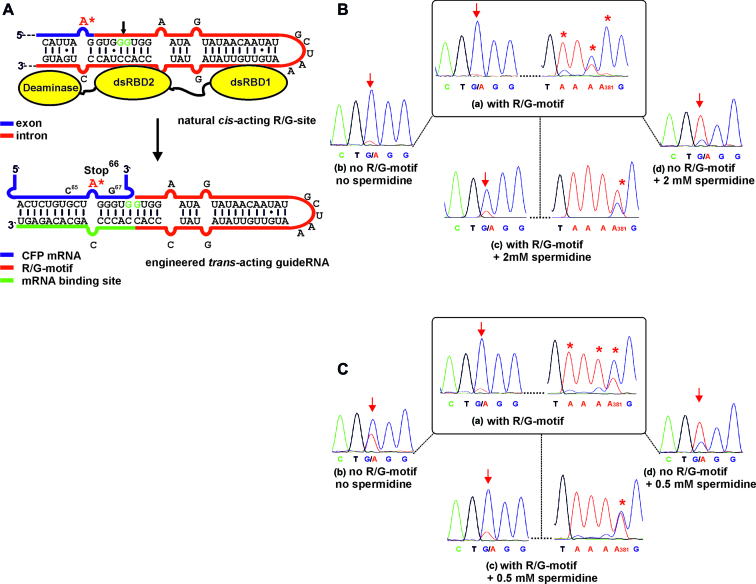
Figure 1.
(A) Engineering the natural cis-acting R/G-site into a trans-acting guideRNA that steers wild-type hADAR2 to a reporter mRNA (CFP) to repair the Stop codon 66 (UA*G) to tryptophan. (B and C) In vitro RNA editing experiments in absence and presence of spermidine and of the R/G-motif in the guideRNA. Targeted adenosine is marked with an arrow; off-site editing around adenosine 381 is marked with *. The sequence of the control guideRNA lacking the R/G-motif was 5΄-NNN GAACACCCC*AGCACAGA. (B) Editing under high concentrations ([ADAR2] = 350 nM, [guideRNA] = 125 nM, [mRNA] = 25 nM, [Mg] = 3 mM). (C) Editing under low concentrations ([ADAR2] = 180 nM, [guideRNA] = 5 nM, [mRNA] = 0.5 nM, [Mg] = 1.5 mM).
Human ADAR2 gives highly efficient editing in vitro
First, we studied the new editing strategy in the polymerase chain reaction (PCR) tube with purified guideRNAs and ADAR2 protein. A reporter mRNA (cyan fluorescent protein) that contains a single G-to-A point mutation generating a nonsense stop signal (Trp66→amber) served as substrate (Figure 1), as described before (34,42). Wild-type human ADAR2 was expressed and purified from yeast (YVH10), as described before (42). To minimize charge repulsion and crowding at the exit of the mRNA, we generated guideRNAs with strictly homogenous 5΄-r(GUGG) ends by in vitro transcription from a hammerhead cassette (44). Initially, we used a guideRNA with a 16 nt reverse complementary mRNA binding site that puts the targeted adenosine in a mismatch with cytosine. The targeted adenosine was kept at a distance of five intervening base pairs to the 5΄-terminus of the R/G-helix and was mismatched with cytosine (Figure 1A).
First, we tested the system at high concentrations of all components ([mRNA] = 25 nM, [guideRNA] = 125 nM, [ADAR2] = 350 nM) in presence of 3 mM magnesium. Full A-to-I conversion at the targeted site (A200) was achieved (Figure 1B, (a)). However, in contrast to our engineered SNAP-ADAR deaminase, (34,42) wild-type ADAR2 was dramatically more reactive and gave massive off-target editing, with full conversion at A381 (Figure 1B, (a)), 50–70% yield at A295, A380, A476 and minor editing at various sites (Supplementary Figure S2). This off-site editing was typically guideRNA-independent. Furthermore, the presence of the R/G-motif was not strictly necessary under these conditions as demonstrated by sufficient editing when applying a 17 nt short, single-stranded guideRNA lacking the R/G-motif (Figure 1B, (b)). Apparently, short RNA duplexes are already sufficiently recognized by ADAR2 under these lax conditions. Consequently, we increased the stringency by adding spermidine (2 mM, Figure 1B, (c and d)). The additive diminished over-editing at all sites to background except adenosine 381 that retained some residual activity (Figure 1B, (c)); Supplementary Figure S3). In presence of spermidine, the guideRNA lacking the R/G-motif was clearly inferior compared to the complete R/G-guideRNA (Figure 1B, (d) versus (c)). We also tested the reverse case and demonstrated that the R/G-guideRNA does not recruit the engineered SNAP-ADAR2 deaminase that lacks both dsRNA binding domains (Supplementary Figure S5). Thus, the recognition between R/G-motif and the dsRBDs is required for editing.
Then we studied editing at low concentrations of the components (Figure 1C). For this, we halved the ADAR2 concentration to 180 nM and strongly decreased the concentrations of the guideRNA (5 nM) and of the mRNA (0.5 nM). Furthermore, the magnesium concentration was decreased to physiological levels (1.5 mM). Notably, the editing reaction run much more smoothly, and significant off-target editing occurred only at adenosine 381 (50% yield, Figure 1C, (a)). The addition of spermidine was only possible to 0.5 mM before losing editing yield at the target, and hence, off-site editing was only slightly reduced (Figure 1C, (c)), Supplementary Figure S4). However, 0.5 mM spermidine improved the necessity for the presence of the R/G-motif in the guideRNA clearly (Figure 1C, (b) versus (d)). Overall, off-target editing is controllable to some extent. Complete inhibition, however, appears difficult in the PCR tube.
Editing is selective and efficient in cell culture
In contrast to our alternative SNAP-tag strategy (29,34,35,37), this new strategy allows the ectopic expression of all components including the guideRNA inside the cell. To demonstrate this, we made use of the U6 promotor that enables expression of uncapped small RNAs with homogenous 5΄-GUGG-termini by RNA polymerase III (45–47). Initially, we kept the successful guideRNA architecture from the in vitro experiment, and put the targeted adenosine 6 nt away from the R/G-motif, and kept the adenosine mismatched with cytosine (Figure 2A). Initially, the mRNA binding template was 16 nt in length. GuideRNAs were delivered on pSilencer U6 hygro plasmids (Life Technologies). ADAR2 and the fluorogenic reporter substrate were provided on pcDNA3.1 vectors under control of the CMV promoter. Due to the better brightness we changed the editing substrate from CFP to eGFP (W58amber).
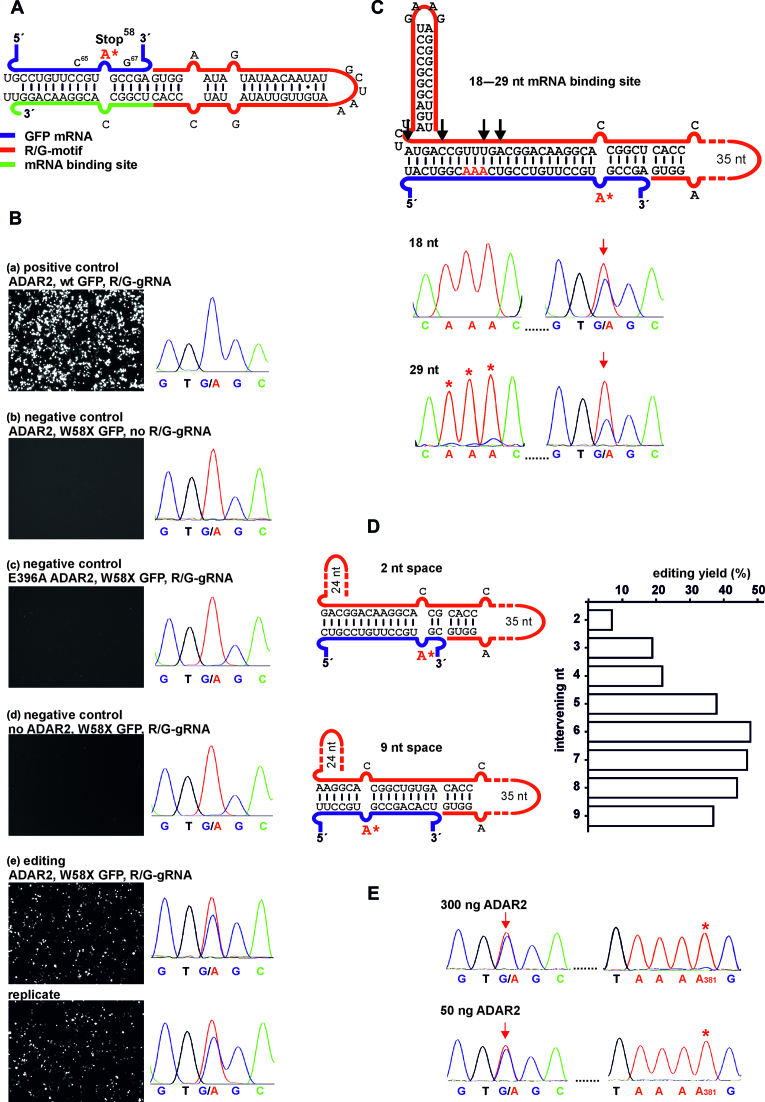
Figure 2.
Editing under transient expression in 293T cells. (A) Scheme of the mRNA/guideRNA duplex of the editing experiment shown in B. (B) Fluorescence imaging of the GFP channel (50 ms exposure time for editing and positive control, 150 ms for negative controls) and RNA sequencing results for cellular editing, 48 h post co-transfection of R/G-guideRNA, ADAR2 and the W58X GFP reporter. E396A ADAR2 is a catalytically inactive mutant. For the editing experiment a duplicate is shown. (C–E) Optimization of cellular editing. (C) Inclusion of a 3΄-terminal hairpin and variation of the length of the mRNA binding platform from 18 to 29 nt, black arrows indicate the different lengths used. For additional traces and fluorescence imaging see Supplementary Figure S10. Red arrows indicate the targeted base, red* indicate off-target editing in the sequencing traces. (D) Editing dependency when varying the number of nucleotides (2–9) intervening the targeted adenosine and the R/G-motif. (E) Editing versus off-target editing using the optimal guideRNA architecture (7 intervening bases, 16 nt mRNA template, 3΄-hairpin) depending on the amount of transfected ADAR2. If not indicated differently, plasmid amounts were 300 ng ADAR2, 300 ng W58X GFP, 1300–1600 ng guideRNA per well in 24-well-plates. For details see Supplementary Figures S8–S15.
After co-transfection of all three plasmids (ADAR2, GFP reporter, guideRNA) into 293T cells the fluorescence phenotype was analyzed via fluorescence microscopy (Figure 2B). Initial optimization showed that a stoichiometry of five guideRNA to one ADAR2 plasmid was optimal. Transfection with wt GFP served as positive control (Figure 2B, (a)). The fluorescence of the Stop58 eGFP transcript was only restored in presence of both components: ADAR2 and a guideRNA reverse complementary to the target site (Figure 2B, (e)). Applying no guideRNA (Figure 2B, (b)) or a guideRNA that binds to the transcript, but 24 nt downstream the target site [not shown], showed no repair activity. This is in accordance with the strict requirement for a dsRNA secondary structure at the editing site. Editing was also absent when a catalytically inactive ADAR2 variant (E396A) or no ADAR2 was used (Figure 2B, (c and d)). We extracted total RNA from the cells, reverse transcribed and amplified the eGFP transcript with specific primers to estimate the editing yield by Sanger sequencing. This revealed a single and specific A-to-G conversion at the targeted codon with approximately 25% yield after 24 h (Supplementary Figure S8) and up to 40% yield after 48 h (Figure 2B, (e)). The Sanger sequencing trace covered the whole coding sequence of the reporter and revealed a highly specific editing. Only at adenosine 381, which was already prone to off-target editing in vitro (Figure 1B and C), we found some minor (<5%) guideRNA-independent off-target editing (full trace see Supplementary Figure S6). However, by adjusting the amount of co-transfected ADAR2 (see next paragraph) this specific off-target editing event was completely abolished.
Further control experiments demonstrated the benefit of the R/G-motif in our guideRNAs. First, when applying our R/G-guideRNA together with SNAP-ADAR2 (lacking the dsRBD that recognize the R/G-motif) instead of human ADAR2 (Supplementary Figure S9), no editing took place. Second, chemically stabilized, single-stranded guideRNAs of 19 nt or 21 nt length that put the targeted adenosine almost centrally into an A:C mismatch but lacks the R/G-motif elicited some minor editing but was clearly inferior to the R/G-guideRNA in recruiting human ADAR2 to the reporter gene (Supplementary Figure S9).
Editing is further improved by optimizing the guideRNA
To further improve cellular editing we pursued several strategies. First, a hairpin, derived from the BoxB-motif (48,49), was included at the 3΄-terminus of the R/G-guideRNA to stabilize it. However, the effect was relatively small. As the hairpin did not show any drawback for the editing yields but facilitated cloning, the hairpin was included in all guideRNA architectures that followed. We then varied the length of the mRNA binding template from 18 to 29 nt with several intermediates (Figure 2C, and Supplementary Figure S10). When using up to 20 nt, the editing yield stayed virtually unchanged around 40%. However, when increasing the mRNA binding site to 25 and 29 nt, the editing yield significantly dropped (Supplementary Figure S10). Furthermore, the risk for off-target editing in the mRNA/guideRNA duplex increased with increasing the guideRNA's length (Figure 2C, Supplementary Figure S10), reminiscent to the situation in the first editing trials by Tod Woolf (33). We then systematically varied the distance between the editing site and the 5΄-terminus of the guideRNA (Figure 2D, Supplementary Figure S11). Notably, we found a bell-shaped distribution of the editing yield. Whereas almost no editing was observed with two intervening bases, the yield increased step-wise and reached an optimum with 6 to 7 intervening bases and decreased slowly for ≥8 nucleotides. Thus, changing the guideRNA architecture from the original 10+1+5 nt design to 8+1+7 nt not only shifts the targeted bases further into the middle of the mRNA/gRNA-duplex but also improves the editing yield from around 40% up to 50% (Figure 2B versus 2D and E). The same bell-shaped trend with a maximum around 7 intervening nucleotides was repeatedly found for the repair of the R407Q missense mutation (5΄-CAG codon) in PINK1 (Supplementary Figure S15). Under transient transfection, editing increased until 48 h (50%), stayed constant until 72 h and then started to decline slowly (40% after 96 h, Supplementary Figure S13). Finally, the amount of ADAR2 in the transfection was optimized to reduce off-target editing. Indeed, when reducing the amount of transfected ADAR2 to a range of 25-200 ng, the off-target editing had disappeared without interfering target editing (Figure 2E, Supplementary Figures S7 and S14).
Editing works efficiently under genomic expression of ADAR2
Unfortunately, editing failed in 293T cells without overexpression of ADAR2 from a plasmid (Figure 2B, (d)). By RT-qPCR we could show that neither ADAR2 nor its typical endogenous target GluR2 is expressed in 293T cells. The plasmid-borne ADAR2 was on average expressed to the ∼11-fold mRNA-level of the reference gene β-actin (Supplementary Figure S16). To better mimic endogenous ADAR2-levels, we created a homogenous 293T cell line containing a single genomic copy of human ADAR2 under control of the CMV tet-on promoter. Under full induction with doxycycline, the mRNA-level of ADAR2 was induced to ∼50% of that of the β-actin gene (Supplementary Figure S16). Into these cells, we transfected the W58X GFP reporter and the optimized guideRNA (7 nt intervening, 3΄-terminal hairpin) delivered on plasmids, as before. Depending on the amount of co-transfected guideRNA (400–1300 ng) editing yields of 45–65% were achieved (Figure 3A, Supplementary Figure S17). No off-target editing was observed in the entire ORF of the reporter gene including adenosine 381 (Figure 3A and Supplementary Figure S18). This is in accordance with the experiments above showing that decreasing ADAR2 expression reduces off-target editing. Notably, the editing yields under lower, but homogenous expression of ADAR2 were significantly better than under transient expression. The advantage of the R/G-motif seems even more pronounced under genomic compared to strong transient ADAR2 expression. The transfection of ADAR2-expressing 293 cells with chemically stabilized, single-stranded guideRNAs of 19 nt and 21 nt length did not elicit detectable editing (Supplementary Figure S9), even though some minor editing was found under transient ADAR2 expression.
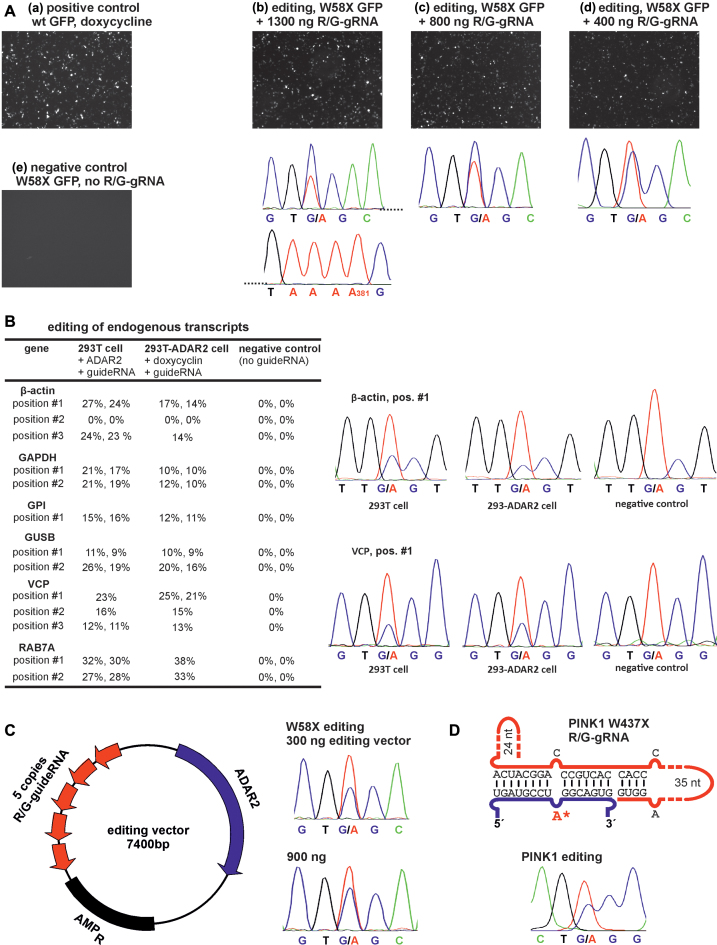
Figure 3.
(A) Editing with genomically integrated ADAR2 in 293T cells under administration of the R/G-guideRNA and the W58X reporter only. (B) Editing of endogenous transcripts. 293T cells were transfected with 300 ng ADAR2 and 1300 ng R/G-gRNA, 293T-ADAR2 cells were induced with 10 ng/ml doxycycline and transfected with 1300 ng guideRNA. Most editing experiments are reported in duplicates. All sequencing traces are given in Supplementary Figure S19. (C) Scheme of the editing vector that combines ADAR2 and 5 identical copies of the guideRNA on a minimal plasmid. Editing of W58X GFP in 239T cells under transient expression shows the functioning. (D) Scheme of the R/G-guideRNA against PINK1 W437amber and editing in 293T cells under transient transfection.
Editing of endogenous transcripts is possible by transfection of the guideRNA only
We then tested the editing of endogenous transcripts, either by co-transfection of the guideRNA with ADAR2 in 293T cells, or by sole transfection of the guideRNA into cells that express ADAR2 under control of doxycycline. Thirteen potentially editable 5΄-UAG triplets in six different genes (β-actin, GAPDH, GPI, GUSB, VCP, RAB7A) were selected. The genes were chosen to cover a range from highly expressed (like β-actin) to lowly expressed (like GUSB). The editing sites were selected not to interfere with gene function, thus they have been located mainly in the 3΄-UTRs. To be able to compare the editing yields, only 5΄-UAG triplets were selected. The guideRNAs were equally designed for all 13 sites following the rules developed above. Twelve out of the 13 sites were editable with yields ranging from 10% to 35% (Figure 3B). With the exception of the VCP and RAB7A transcript, the editing yields seem to benefit from the 20-fold higher ADAR2-level under transient compared to genomic ADAR2 expression. In contrast, the editing yields did not depend on the transcript-levels. In β-actin, for instance, two sites were edited well, whereas one site was not edited at all. On the other hand GUSB, which is roughly 100-fold less expressed than β-actin, was edited to a similar extent as β-actin and with the same trends for transient versus genomic ADAR2 expression. Thus, other factors may determine the editing success like the accessibility of the edited site.
Guided RNA editing repairs the PINK1 W437amber mutation and rescues mitophagy
To demonstrate the practical application we aimed to repair a disease-causing point mutation under rescue of the disease-relevant phenotype. The PINK1 W437Stop mutation was selected as it is linked to an inheritable monogenetic form of Parkinson's disease (40) and is characterized by a well-known cellular phenotype (loss of mitophagy). Assuming that all three Stop codons at position 437 in PINK1 are disease-relevant, we chose the amber codon, which is the best editable (42). Following the rules developed above, a guideRNA was designed that puts the amber Stop codon at position 437 into an A:C mismatch in the middle of a 16 nt mRNA/guideRNA duplex (Figure 3D). The guideRNA was further stabilized by a hairpin at the 3΄-terminus. Co-transfection of plasmids encoding PINK1 W437amber, ADAR2 and the guideRNA in 293T cells resulted in an editing yield of 35% (Figure 3D).
The established PINK1 functional assay in HeLa cells follows a complex protocol (41). First, endogenous wild-type PINK1 is knocked down by RNAi. Then mutated PINK1 and eGFP-tagged Parkin are co-transfected on plasmids, and the mitochondrial membrane potential of the cells is depolarized by CCCP treatment. The CCCP-induced PINK1-dependent perinuclear clustering of Parkin is typically visible after 2 h of CCCP treatment. After long-term treatment (24 h CCCP) mitochondria are cleared from the cytoplasm (mitophagy). To facilitate the protocol and reduce the number of transfections, we simplified the assay. First, a stable PINK1 knock out (KO) HeLa cell line was created using CRISPR-Cas9 (50) technology (for details see Supplementary Figure S20). Second, an editing vector was created that contains ADAR2-His6 together with five copies of the PINK1-guideRNA, each under control of a U6-promotor (Figure 3C). This allowed us to apply a triple (PINK1, Parkin, editing vector) instead of a quadruple (PINK1, Parkin, ADAR2, guideRNA) co-transfection. Furthermore, the editing vector ensures that guideRNA and ADAR2 are taken-up in a defined stoichiometry and allows to deduce guideRNA expression from ADAR2 staining. The size of the vector was reduced to 7.4 kbp by removing all unnecessary parts of the backbone. A similar construct that targets the W58X GFP reporter was functional in 293T cells, however, suffering from slightly reduced editing yields (35% instead of 50%).
As shown in Figure 4A/B, (a), wild-type HeLa cells show the typical perinuclear clustering of Parkin after 2 h CCCP treatment in 89 ± 3% of the Parkin-positive cells. This clustering phenotype has been shown repeatedly (41,51) to require functional PINK1. As expected, Parkin-clustering disappeared in the PINK1-KO HeLa cell line, even when W437amber PINK1 was overexpressed from a plasmid (Figure 4A/B, (b)), clearly showing that W437amber is unable to rescue the phenotype. However, Parkin-clustering was fully rescued by co-transfection of PINK1 W437X together with the editing vector in 85±8% of the cells positive for PINK1, Parkin and ADAR2 expression. Co-transfection of PINK1amber/Parkin with the guideRNA or with ADAR2 alone (Figure 4A/B, (d and e)) was unable to rescue the clustering phenotype, even though some clustering has been found occasionally in a low number of the cells (≤15%). Consequently, the presence of both components of the editing system, the guideRNA and ADAR2, are required to rescue the phenotype to the level seen for the wild-type control. To asses editing efficiency, total RNA was extracted from cells, reverse transcribed with PINK1-specific primers and was Sanger-sequenced after PCR amplification. Only in presence of both components, guideRNA and ADAR2, editing of the PINK1 W437amber codon was detectable (ca. 10% yield). The formation of full-length PINK1 after editing was further confirmed by Western blot (Supplementary Figure S21). To ensure that the rescue of Parkin-clustering was due to the repair of the PINK1 W437amber codon, we co-transfected HeLa KO cells with Parkin and the editing vector, but in absence of the editing target PINK1 W437X. As expected no rescue was detectable (Figure 4A, (f)). Obviously, restoring the loss-of-function phenotype was only mediated by site-directed RNA editing of the amber Stop codon of the transfected PINK1 W437X construct. Finally, we provide data for the PINK1/Parkin-mediated mitophagy, which is restored after editing of PINK1 W437amber (Figure 4A). For this, the mitochondria of all cells were stained after 0 h, 2 h and 24 h of CCCP with Mito-Tracker (Red CMXRos). As before, after 2 h of CCCP treatment Parkin did clearly co-localize with the perinuclear mitochondria. After 24 h of CCCP treatment, mitochondrial staining has disappeared in PINK1/Parkin/ADAR2 triple-positive cells but not in cells that lack the expression of one or several of the transfected components (Figure 4C, Supplementary Figure S23). This confirms that the two stages of mitophagy, (41,51) the Parkin-clustering (after 2 h of depolarization) and the clearance of depolarized mitochondria (after 24 h of depolarization), are restored upon editing of PINK1 W437amber.
CONCLUSION
We demonstrate here the first strategy to harness wild-type human ADAR2 to stimulate site-selective RNA editing at arbitrary mRNAs. The strategy relies on the ectopic expression of short, structured guideRNAs that base-pair to specific, user-defined mRNAs thereby mimicking the intronic R/G-motif of the glutamate receptor transcript to recruit wild-type human ADAR2 to stimulate A-to-I conversion. In the PCR tube, we demonstrated the functionality of the tool and the dependency on the structured R/G-motif. Selective and efficient editing was achieved even though the tendency for off-site editing required optimization of the reaction conditions and was not entirely controllable. As the guideRNAs can be expressed from an RNA polymerase III promoter, the strategy is fully genetically encodable. We have demonstrated the specific and efficient (up to 40%) editing of a fluorescent reporter gene in 293T cells. Editing benefited from the presence of the structured R/G-motif in the guideRNA, and the transfection of single-stranded, chemically stabilized guideRNAs (with a binding site for the target mRNA up to 21 nt) were not as efficient as our guideRNAs with a 16 nt mRNA binding site plus the additional R/G-motif. In contrast to in vitro editing, no off-target editing was observed in the reporter gene within cells once the amount of ADAR2 plasmid was optimized. The lower amount of ADAR2 and the RNP-landscape inside the living cell possibly protects the transcript from over-editing without suppressing editing at the targeted adenosine. Editing was further improved to a yield of ∼50% by optimizing the guideRNA architecture. In particular the placement of the targeted adenosine with respect to the R/G-motif was critical. The optimal architecture turned out to deviate slightly from the natural R/G-site of GluR2. The insertion of additional nucleotides seems reasonable to accommodate the exit of the mRNA's 3΄-part (28,43). Site-directed RNA editing worked efficiently in a created 293T cell line that expresses human ADAR2 from the genome under the control of doxycycline. Even though ADAR2 mRNA-levels were >20-fold reduced compared to transient expression conditions, improved editing yields (up to 65%) were achieved. Furthermore, off-site editing in the reporter was abolished and the amount of R/G-guideRNA could be reduced. Finally, we demonstrated with a set of 13 sites in six housekeeping genes that ADAR2 can be re-directed for the editing of endogenously expressed transcripts. Importantly, this was also successful by ectopic expression of the guideRNA alone into the engineered 293 cells that express ADAR2 moderately from a single genomic copy. However, as the guideRNAs have not been optimized for these targets, editing yields stayed in a range of 10–35%. Further optimization of the guideRNAs may focus on the R/G-motif, the mRNA binding-site, and chemical modifications (29). In this work, we have mostly focused on the editing of 5΄-UAG triplets. We expect many other triplets to be well editable, (42) an example for 5΄-CAG is shown in the Supporting Information (Supplementary Figure S15). However, one can expect different triplets to be unequally well edited corresponding to the well-known preferences of the deaminase domain (52).
To demonstrate the usefulness of the editing approach in biological applications, we generated an editing vector that contains one copy of human ADAR2 and five copies of the R/G-guideRNA. This vector allowed us to define the stoichiometry of ADAR2 and the R/G-guideRNA in the transfected cell and to strongly reduce the amount of plasmid. We showed the functioning of the vector for the repair of the W58X GFP reporter. The respective editing vector against PINK1 W437X was functional to rescue the PINK1-mediated perinuclear clustering of Parkin and the mitophagy phenotype in PINK1-KO HeLa cells under transient transfection of the editing target. We could clearly demonstrate the rescue to require the presence of the entire editing machinery, the guideRNA and ADAR2, as well as the presence of the editing target PINK1 W437X. Editing was specific and no other A-to-I editing event was detectable in the Sanger sequencing trace, which covered ca. 600 nt (140 adenosines) of the PINK1 transcript (Supplementary Figure S22). However, to assess how the guide-RNA-dependent harnessing of ADAR2 affects the editing homeostasis at natural editing sites, transcriptome-wide deep RNA-sequencing would be required, preferentially in an in vivo situation where endogenous ADAR2 function is detectable and required. The editing yield of 10% for PINK1 W437X can surely be further improved, for instance by optimizing the co-transfection conditions in HeLa cells. The apparent editing yield over the entire cell culture suffers from the expression of PINK1 in the absence of the editing components. Thus, we expect an editing yield higher than 10% in those cells that actually express the editing machinery. Anyway, the rescue of this recessive loss-of-function mutation in PINK1 may indeed only require a relatively moderate repair yield.
Together, we have shown that harnessing human ADAR2 for site-directed RNA editing allows recoding mRNAs to levels high enough to manipulate disease-relevant cellular phenotypes. Other endogenous RNA-processing enzymes, including RNaseH and the RNA-induced silencing complex (RNA interference), have been shown to be re-addressable toward new targets by ectopic expression or administration of short (chemically stabilized) guideRNAs (53,54). The structured R/G-guideRNAs introduced here are capable of recruiting human ADAR2. It remains yet unclear if they are able to recruit endogenously expressed ADAR2 for site-directed RNA editing. Certainly, they are a good starting point for the development of improved guideRNA architectures in the future. While RNAi and RNaseH recruitment are limited to the up/down-regulation of target transcripts, RNA editing allows for the active recoding and hence enables an entirely novel point of attack. Currently, there is increasing success in the tailoring of oligonucleotide drugs with respect to their efficacy, stability, toxicology and delivery (55). Major breakthrough seems to have happened in the therapeutic RNA interference field currently (31,56,57). We feel confident that our work sets the stage for the development of a novel therapeutic platform that is based on the harnessing of human ADARs for the repair of disease-relevant genes by either ectopic expression or administration of chemically stabilized (29) guideRNAs.
Supplementary Material
ACKNOWLEDGEMENTS
The CRISPR/Cas9 plasmid (pSpCas9(PINK1-KO)-2A-Puro) was generously provided by Christine Bus (University of Tübingen, Germany).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Tübingen, the Hertie Foundation, the German Center for Neurodegenerative Disease and the Deutsche Forschungsgemeinschaft [STA 1053/3-2 to T.S., GE 2844/1-1 to S.G.); European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program [grant no. 647328]. Funding for open access charge: DFG.
Conflict of interest statement. The Authors have filed a patent on the technology described in this article.
REFERENCES
- 1.Nishikura K. Functions and Regulation of RNA Editing by ADAR Deaminases. Annu. Rev. Biochem. 2010; 79:321–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K.. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007; 315:1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K.. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006; 13:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rueter S.M., Dawson T.R., Emeson R.B.. Regulation of alternative splicing by RNA editing. Nature. 1999; 399:75–80. [DOI] [PubMed] [Google Scholar]
- 5.Rueter S.M., Burns C.M., Coode S.A., Mookherjee P., Emeson R.B.. Glutamate receptor RNA editing in vitro by enzymatic conversion of adenosine to inosine. Science. 1995; 267:1491–1494. [DOI] [PubMed] [Google Scholar]
- 6.Melcher T., Maas S., Herb A., Sprengel R., Seeburg P.H., Higuchi M.. A mammalian RNA editing enzyme. Nature. 1996; 379:460–464. [DOI] [PubMed] [Google Scholar]
- 7.Bass B.L. RNA editing by Adenosine Deaminases that act on RNA. Annu. Rev. Biochem. 2002; 71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morabito M.V., Abbas A.I., Hood J.L., Kesterson R.A., Jacobs M.M., Kump D.S., Hachey D.L., Roth B.L., Emeson R.B.. Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader-Willi syndrome. Neurobiol. Dis. 2010; 39:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maas S., Kawahara Y., Tamburro K.M., Nishikura K.. A-to-I RNA editing and human disease. RNA Biol. 2006; 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slotkin W., Nishikura K.. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013; 5:105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silberberg G., Lundin D., Navon R., Öhman M.. Deregulation of the A-to-I RNA editing mechanism in psychiatric disorders. Hum. Mol. Genet. 2012; 21:311–321. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H.. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000; 406:78–81. [DOI] [PubMed] [Google Scholar]
- 13.Rice G.I., Kasher P.R., Forte G.M., Mannion N.M., Greenwood S.M., Szynkiewicz M., Dickerson J.E., Bhaskar S.S., Zampini M., Briggs T.A. et al. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 2012; 44:1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X.J., He P.P., Li M., He C.D., Yan K.L., Cui Y., Yang S., Zhang K.Y., Gao M., Chen J.J. et al. Seven novel mutations of the ADAR gene in Chinese families and sporadic patients with dyschromatosis symmetrica hereditaria (DSH). Hum. Mutat. 2004; 23:629–630. [DOI] [PubMed] [Google Scholar]
- 15.Hartner J.C., Schmittwolf C., Kispert A., Müller A.M., Higuchi M., Seeburg P.H.. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004; 279:4894–4902. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q., Miyakoda M., Yang W., Khillan J., Stachura D.L., Weiss M.J., Nishikura K.. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004; 279:4952–4961. [DOI] [PubMed] [Google Scholar]
- 17.Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C., Li J.B., Seeburg P.H., Walkley C.R.. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015; 349:1115–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mannion N.M., Greenwood S.M., Young R., Cox S., Brindle J., Read D., Nellaker C., Vesely C., Ponting C.P., McLaughlin P.J. et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014; 9:1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L., Li Y., Lin C.H., Chan T.H., Chow R.K., Song Y., Liu M., Yuan Y.F., Fu L., Kong K.L. et al. Recoding RNA editing of antizyme inhibitor 1 predisposes to hepatocellular carcinoma. Nat. Med. 2013; 19:209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimokawa T., Rahman M.F., Tostar U., Sonkoly E., Ståhle M., Pivarcsi A., Palaniswamy R., Zaphiropoulos P.G.. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biol. 2013; 10:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallo A. RNA editing enters the limelight in cancer. Nat. Med. 2013; 19:130–131. [DOI] [PubMed] [Google Scholar]
- 22.Paz-Yaacov N., Bazak L., Buchumenski I., Porath H.T., Danan-Gotthold M., Knisbacher B.A., Eisenberg E., Levanon E.Y.. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Reports. 2015; 13:267–276. [DOI] [PubMed] [Google Scholar]
- 23.Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M., Eterovic A.K., Yuan Y., Li J. et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015; 28:515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picardi E., Manzari C., Mastropasqua F., Aiello I., D'Erchia A.M., Pesole G.. Profiling RNA editing in human tissues: Towards the inosinome Atlas. Sci. Rep. 2015; 5:14941–14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng Z., Cheng Y., Tan B.C., Kang L., Tian Z., Zhu Y., Zhang W., Liang Y., Hu X., Tan X. et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotech. 2012; 30:253–260. [DOI] [PubMed] [Google Scholar]
- 26.Bazak L., Haviv A., Barak M., Jacob-Hirsch J., Deng P., Zhang R., Isaacs F.J., Rechavi G., Li J.B., Eisenberg E. et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014; 24:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramaswami G., Lin W., Piskol R., Tan M.H., Davis C., Li J.B.. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Meth. 2012; 9:579–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefl R., Oberstrass F.C., Hood J.L., Jourdan M., Zimmermann M., Skrisovska L., Maris C., Peng L., Hofr C., Emeson R.B. et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010; 143:225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogel P., Stafforst T.. Site-directed RNA editing with Antagomir Deaminases — A tool to study protein and RNA function. ChemMedChem. 2014; 9:2021–2025. [DOI] [PubMed] [Google Scholar]
- 30.Cox D.B., Platt R.J., Zhang F.. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015; 21:121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sehgal A., Barros S., Ivanciu L., Cooley B., Qin J., Racie T., Hettinger J., Carioto M., Jiang Y., Brodsky J. et al. An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat. Med. 2015; 21:491–497. [DOI] [PubMed] [Google Scholar]
- 32.Stern M., Alton E.W.F.W.. Albeda SM. Use of liposomes in the treatment of cystic fibrosis. Gene Therapy in Lung Disease: Lung Biology in Health and Disease. 2002; 169, NY: CRC Press; 383–396. [Google Scholar]
- 33.Woolf T.M., Chase J.M., Stinchcomb D.T.. Towards the therapeutic editing of mutated RNA sequences. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:8298–8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stafforst T., Schneider M.F.. An RNA–Deaminase conjugate selectively repairs point mutations. Angew. Chem. Int. Ed. Engl. 2012; 51:11166–11169. [DOI] [PubMed] [Google Scholar]
- 35.Vogel P., Schneider M.F., Wettengel J., Stafforst T.. Improving site-directed RNA editing in vitro and in cell culture by chemical modification of the guideRNA. Angew. Chem. Int. Ed. Engl. 2014; 53:6267–6271. [DOI] [PubMed] [Google Scholar]
- 36.Montiel-Gonzalez M. F., Guillermo I., Yudowski A., Rosenthal J. J. C.. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:18285–18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanswillemenke A., Kuzdere T., Vogel P., Jékely G., Stafforst T.. Site-directed RNA editing in vivo can be triggered by the light-driven assembly of an artificial riboprotein. J. Am. Chem. Soc. 2015; 137:15875–15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization (WHO). Neurological disorders: Public health challenges. 2006; Geneva; ISBN 9241563362. [Google Scholar]
- 39.Farrer M.J. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat. Rev. Gen. 2006; 7:306–318. [DOI] [PubMed] [Google Scholar]
- 40.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G. et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004; 304:1158–1160. [DOI] [PubMed] [Google Scholar]
- 41.Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W.. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010; 12:119–131. [DOI] [PubMed] [Google Scholar]
- 42.Schneider M.F., Wettengel J., Hoffmann P.C., Stafforst T.. Optimal guideRNAs for re-directing deaminase activity of hADAR1 and hADAR2 in trans. Nucleic Acids Res. 2014; 42:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthews M.M., Thomas J.M., Zheng Y., Tran K., Phelps K.J., Scott A.I., Havel J., Fisher A.J., Beal P.A.. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat. Struct. Mol. Biol. 2016; 23:426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Persson T., Hartmann R. K., Eckstein F.. Selection of hammerhead ribozyme variants with low Mg2+ requirement: importance of stem-loop II. ChemBioChem. 2002; 3:1066–1071. [DOI] [PubMed] [Google Scholar]
- 45.Miyagishi M., Taira K.. U6 promoter-driven siRNAs with four uridine 3΄ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotech. 2002; 20:497–500. [DOI] [PubMed] [Google Scholar]
- 46.Lee N. S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J.. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotech. 2002; 20:500–505. [DOI] [PubMed] [Google Scholar]
- 47.Paul C. P., Good P. D., Winer I., Engelke D. R.. Effective expression of small interfering RNA in human cells. Nat. Biotech. 2002; 20:505–508. [DOI] [PubMed] [Google Scholar]
- 48.Daigle N., Ellenberg J.. LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat. Methods. 2007; 4:633–636. [DOI] [PubMed] [Google Scholar]
- 49.Keryer-Bibens C., Barreau C., Osborne H. B.. Tethering of proteins to RNAs by bacteriophage proteins. Biol. Cell. 2008; 100:125–138. [DOI] [PubMed] [Google Scholar]
- 50.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S. et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eggington J.M., Greene T., Bass B.L.. Predicting sites of ADAR editing in double-stranded RNA. Nat. Commun. 2011; 2, doi:10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett C.F., Swayze E.E.. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010; 50:259–293. [DOI] [PubMed] [Google Scholar]
- 54.Kole R., Krainer A.R., Altman S.. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug. Discov. 2012; 11:125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reautschnig P., Vogel P., Stafforst T.. The notorious R.N.A. in the spotlight – drug or target for the treatment of disease. RNA Biol. 2016; doi:10.1080/15476286.2016.1208323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E. et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013; 369:819–829. [DOI] [PubMed] [Google Scholar]
- 57.Fitzgerald K., Frank-Kamenetsky M., Shulga-Morskaya S., Liebow A., Bettencourt B.R., Sutherland J.E., Hutabarat R.M., Clausen V.A., Karsten V., Cehelsky J. et al. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: A randomised, single-blind, placebo-controlled, phase 1 trial. Lancet. 2014; 383:60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.