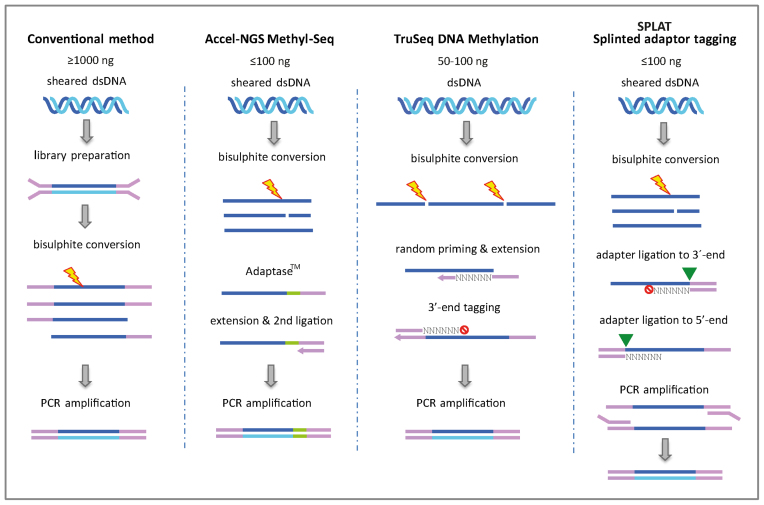
Figure 1.
Principles of library preparation methods for whole genome bisulphite sequencing. In the conventional workflow (MethylC-seq) methylated adapters are ligated to double stranded sheared DNA fragments. The constructs are then bisulphite converted prior to amplification with a uracil reading PCR polymerase. The Accel-NGS Methyl-Seq uses the proprietary Adaptase™ technology to attach a low complexity sequence tail to the 3΄-termini of pre-sheared and bisulphite-converted DNA, and an adapter sequence. After an extension step a second adapter is ligated and the libraries are PCR amplified. The TruSeq DNA Methylation method (formerly EpiGnome) uses random hexamer tagged oligonucleotides to simultaneously copy the bisulphite-converted strand and add a 5΄-terminal adaptor sequence. In a subsequent step, a 3΄-terminal adapter is tagged, also by using a random sequence oligonucleotide. In the SPLAT protocol adapters with a protruding random hexamer are annealed to the 3΄-termini of the single stranded DNA. The random hexamer acts as a ‘splint’ and the adapter sequence is ligated to the 3΄-termini of single stranded DNA using standard T4 DNA ligation. A modification of the last 3΄- residue of the random hexamer is required to prevent self-ligation of the adapter. In a second step, adapters with a 5΄-terminal random hexamer overhang is annealed to ligate the 5΄-termini of the single stranded DNA, also using T4 DNA ligase. Finally the SPLAT libraries are PCR amplified using a uracil reading polymerase.