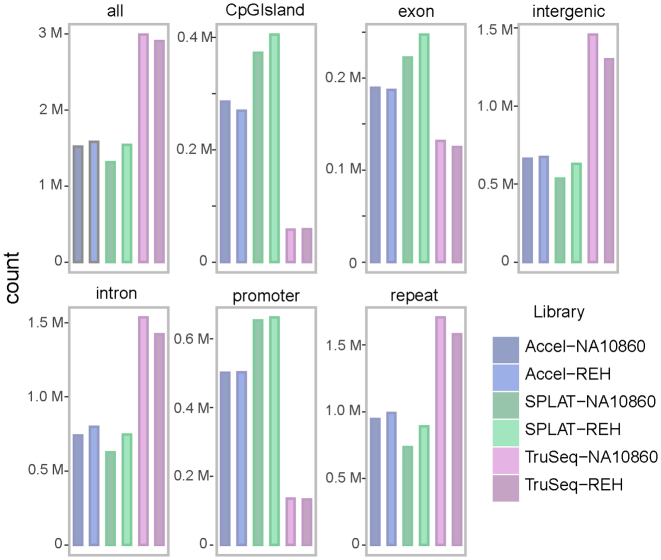
Figure 6.
Annotation of CpG sites with low coverage in the ‘post bisulphite’ WGBS methods. A coverage threshold of ≤2 reads was applied to identify CpG sites that were insufficiently represented by the different WGBS library methods (based on down-sampled data). In TruSeq DNA Methylation libraries, low coverage sites were mostly found in intergenic regions and introns. The SPLAT and Accel-NGS Methyl-Seq data displayed overall lower number of CpG sites with low coverage, although the number of poorly represented sites in CpG islands and promoter regions were higher compared to the TruSeq DNA methylation libraries.