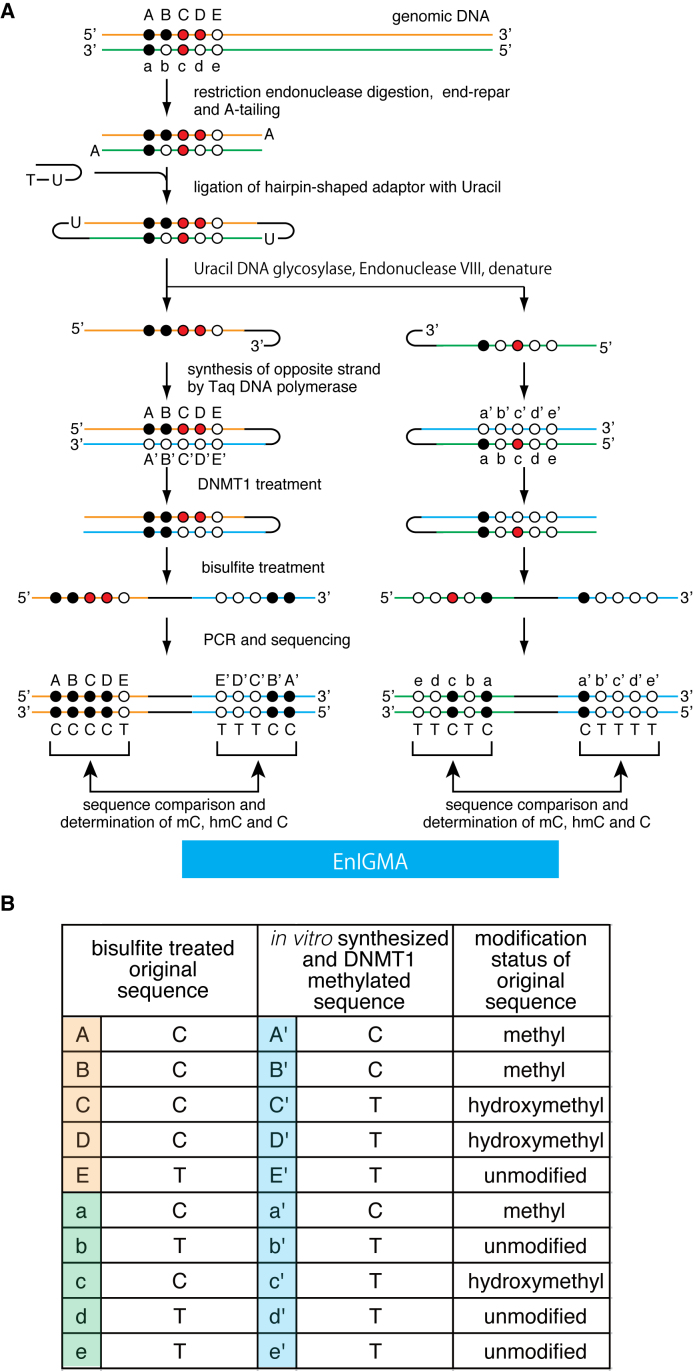
Figure 1.
(A) A schematic of the EnIGMA method is shown. The black circles designate methylated cytosine, the red circles hydroxylmethylated cytosine and the white circles unmodified cytosine. The EnIGMA method analyzes the CpGs on one strand (orange line for top strand and green line for bottom strand in this figure). The DNA is digested by appropriate restriction endonuclease, and the hairpin DNA is ligated. Next the opposite strand DNA is synthesized in vitro (blue line). Resulted DNA is methylated by DNMT1 enzyme followed by bisulfite conversion and PCR by specific primers. (B) The decoding table for cytosine modification status of the CpGs shown in (A).