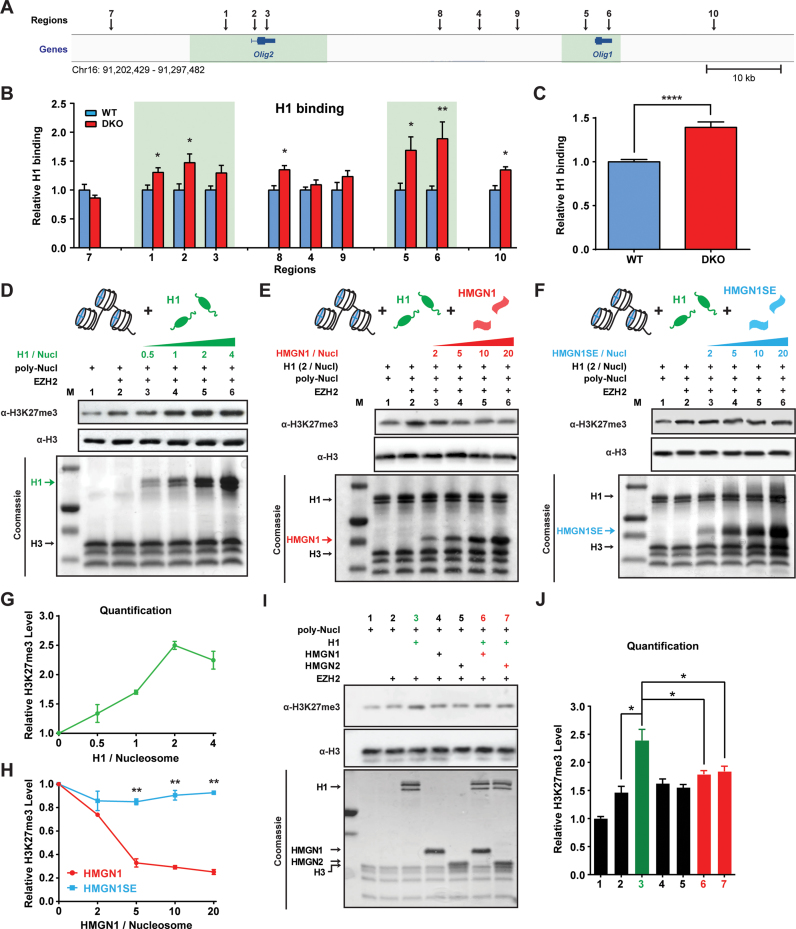
Figure 4.
HMGN histone H1 interplay modulates EZH2-mediated H3K27me3. (A) Genome browser snapshot shows 10 regions (indicated by arrows) near Olig1&2 genes analyzed for histone H1 binding. Sites 1–6 correspond to the same sites in Figure 3. (B) Relative enrichment in H1 occupancy at the indicated sites in DKO cells, normalized to occupancy in WT cells. Data were obtained from two ESC clones (biological replicates) in two independent experiments and presented as mean ± SEM. Primers used are listed in Supplementary Table S1. (C) Average increase in H1 occupancy at the selected sites in DKO chromatin. (D) Histone methyl transfer (HMT) assays, using H1-depleted poly-nucleosomes (poly-Nucl) as substrates show H1 dose dependent increase in EZH2 mediated H3K27me3 levels (E and F) HMT assays show that HMGN1 but not HMGN1S20,24E inhibit the H1 mediated enhancement of H3K27me3 levels (G and H) Quantification of panels D–F, the levels of H3K27me3 normalized to H3. (I) HMT assays showing effect of H1, HMGN1 and HMGN2 on H3K27me3 levels. (J) Quantification of panel I. All HMT quantification data were obtained from two independent experiments and presented as mean ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001.