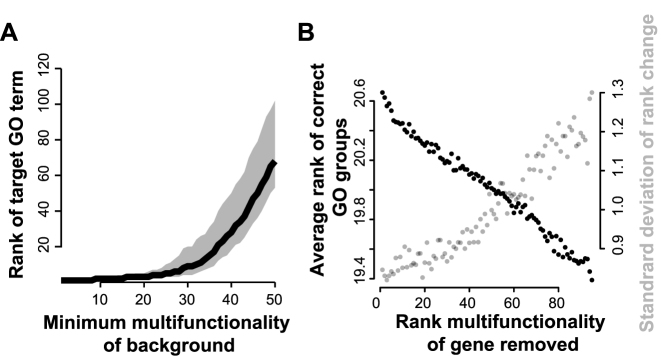
Figure 4.
Simulations showing that the presence of multifunctional genes degrades recovery of ‘true’ functions. (A) Analysis of a simple simulation, in which hit lists of size 100 are generated using 10 genes from a randomly selected GO group (which becomes the target) plus background noise of 90 randomly chosen genes, with the background constrained to have a minimum multifunctionality level. Increasing the minimum multifunctionality of the background genes (x-axis) decreases recovery of the target GO term. Black line indicates average of 1000 simulations; grey area covers 50% of simulations (middle quartiles). (B) Analysis of a more complex model, mixing 10 genes from 10 GO groups to make an artificial hit list, and testing the effect of removing each gene (one at a time, not cumulatively). We plotted the average rank of the target functions against the multifunctionality rank of a single gene removed. Because only a single gene is removed, the effect is modest, but the more multifunctional the gene, the more removing it improves the score but also the more the scores vary. Note the y-axis only includes the range from 19.3–20.7. The plot is the average of 1000 simulations, each of which involved removing each of the 100 genes in turn.