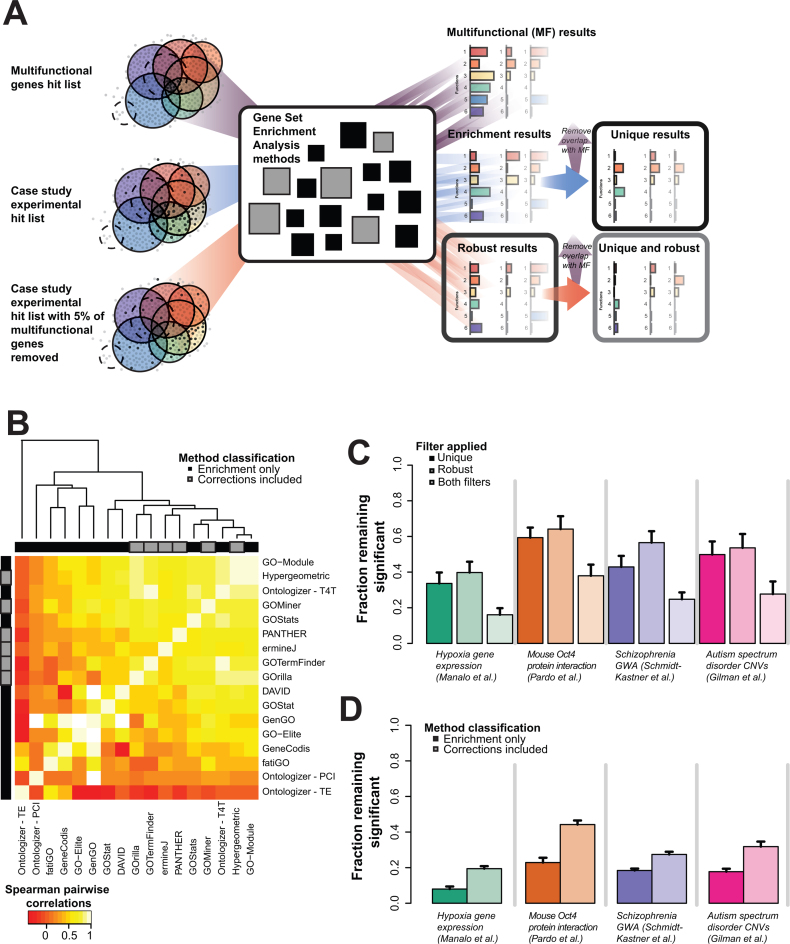
Figure 5.
Effects of multifunctionality on algorithm behavior. (A) Schematic of method to assess uniqueness and robustness of the 17 gene set enrichment methods. We input the top 100 multifunctional genes, the case study genes and then the case study genes filtered at 5% for the most multifunctional genes. The 5% reduced output results are those robust to multifunctionality. The results filtered by the multifunctional results are those used in uniqueness test. (B) The top 100 multifunctional genes were given as a hit list for the individual algorithms, and the resulting GO enrichment results for each were compared. The methods that do not claim to correct cluster together. The corrections that prune results post-enrichment cluster with the non-correcting methods. (C) Four case studies were assessed in each of 17 commonly used enrichment methods and their results assessed for the role of multifunctional genes in generating their systemic results. Only a modest fraction (average 46.4%, average SE 6%) of the reportedly enriched functions are not the same ones that each algorithm outputs when the 100 most multifunctional genes are used as an input (leftmost panels). Removing the 5% most multifunctional genes from each hit list (as few as 1 gene) dramatically alters most reported enrichment, leaving only ∼53.5% (average SE 7%) of them intact (middle panel). This combination of effects has an impact on all but a small fraction (average 26.6%, average SE 5%) of the algorithms across all four studies (right most panels). Note that colors associated with the study are indicated in the legend in panel B. (D) Algorithm behavior is examined for the effects of corrections. We partitioned the algorithms into two classes, those which perform more standard statistical tests (darker colors) and those which attempt to correct for problems with enrichment in some way (lighter colors). We then repeated the analysis from part A. Algorithms attempting to correct their output yield a significantly higher fraction of terms which are both specific (not multifunctional) and robust (to removal of 5% of genes from the hit list).