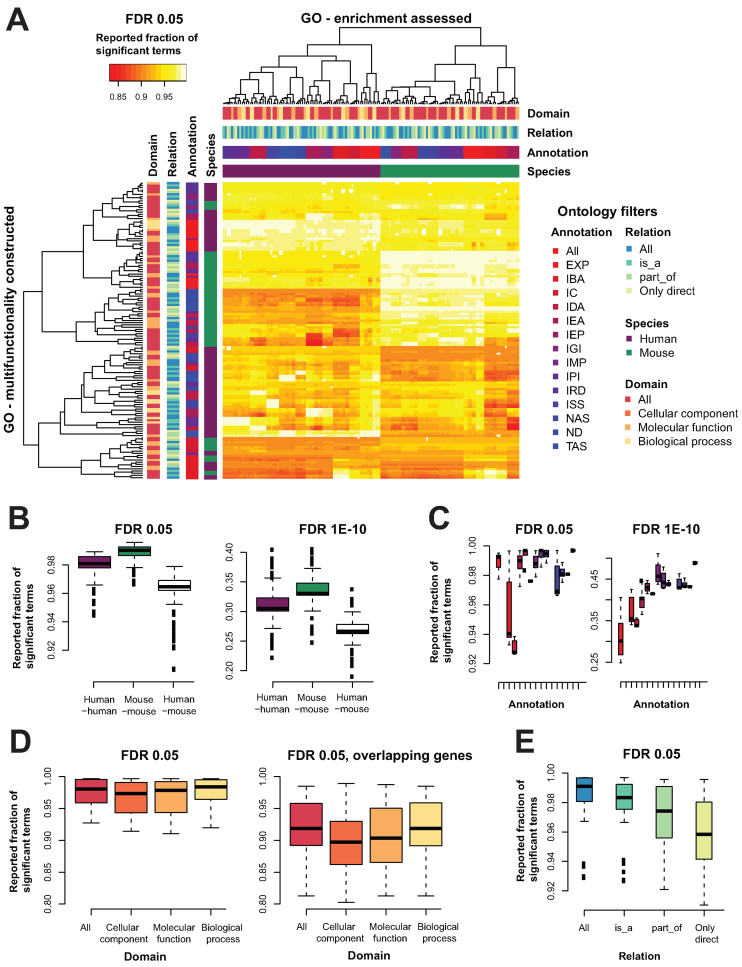
Figure 6.
Multifunctionality bias is a robust feature of GO. Different versions of GO, filtered for different annotation properties were built and compared. (A) Heatmap of the fraction of GO terms enriched in a list of genes ordered by multifunctionality estimated from a different version of GO at an FDR of 0.05. We see a clear structure within a given ontology, with clustering by species (mouse in green, human in purple), then by annotation (shades from red to blue), then by domain (shades from orange to yellow) and some clustering by the relation (shades from blue to green). The color codes listed in the key are used consistently throughout the rest of the panels. (B) The fractions for the original human GO on all the other human GO derivatives, the original mouse GO on all the mouse derivatives, and all the human GOs on the mouse GOs. This is shown for both the FDR < 0.05 and FDR < 1E-10. (C) Taking each multifunctionality list and calculating the enrichment on ‘itself’, we see for the GO's conditioned on annotation codes follow an upward trend to be more enriched as the annotations are more reliable. (D) For domains, this is fairly stable. (E) For relations, it is the opposite, as we become more strict, we lose the multifunctionality bias, moderately.