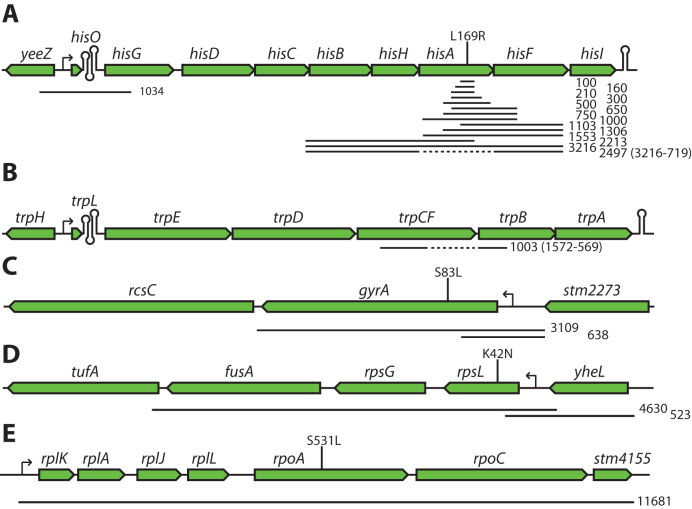
Figure 2.
Examples of Dup-Ins constructed in S. enterica. Black lines below the gene maps indicate sequences duplicated in the different Dup-In constructs. Numbers next to the lines indicate sizes of the duplicated sequences in base pairs. Dashed lines indicate sequences that are missing in deletion mutants. Vertical lines with text above indicate the position of mutations. (A) Fourteen Dup-Ins in the his-operon. The ΔhisA mutant is missing the entire hisA coding sequence (719 bp). Oligos 1–5 (Supplementary Data) were used with different combinations of oligos 6–11 to generate all variants, except the hisO Dup-In for which oligos 12 and 13 were used. (B) Dup-In in the trp operon. The ΔtrpF mutant is missing the last 569 bp of the fused trpCF gene (oligos 14 and 15, Supplementary Data). (C) Two Dup-Ins in the gyrA region (oligo 18 in combination with oligos 16 or 17, Supplementary Data). (D) Two Dup-Ins in the str operon, containing rpsL (oligos 19 and 20, or 21 and 22, Supplementary Data). (E) Dup-In in the alpha operon, containing the rpoB gene (oligos 23 and 24, Supplementary Data).