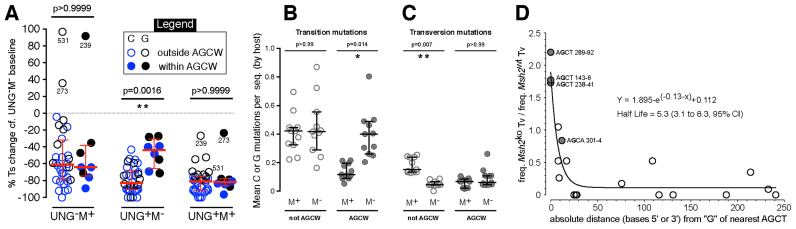
Figure 4.
Processing of deaminations outside versus within AGCW motifs. (A) The change in transition mutation in the top strand at (blue) C or (black) G in each dataset, relative to the baseline UNG–M– dataset, for every site that was mutated more than seven times in the UNG–M– dataset. Medians and interquartile ranges are indicated in red. Significant differences between (solid symbols) AGCW and (open symbols) non-AGCW sites are indicated by Kruskal-Wallis p-values. (B and C) Mean frequencies per sequence of (B) transition mutations or (C) transversion mutations at C:G base pairs lying (open symbols) outside or (closed symbols) inside AGCW sites, for individual hosts of (M+) UNG+M+ or (M–) UNG+M– cells. Bars indicate medians and inter-quartile ranges of hosts means. Significant differences between UNG+M+ cells and UNG+M– cells are indicated by Kruskall-Wallis p-values. (D) The MMR-dependence of normalised transversion mutation at G (y-axis) upon distance from ‘G’ in the nearest AGCT motif (x-axis). The best-fitting exponential decay curve (Prism software) is overlaid.